- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Plasmid DNA Purification Using Filterprep With an Optional Endotoxin Removal Step
Published: Vol 15, Iss 24, Dec 20, 2025 DOI: 10.21769/BioProtoc.5547 Views: 955
Reviewed by: Hemant Kumar PrajapatiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Optimized Protocol for DNA Extraction in Three Theobroma Species
Angie F. Riascos-España [...] Pedro A. Velasquez-Vasconez
May 5, 2025 2020 Views
High-Throughput Indirect Monitoring of TORC1 Activation Using the pTOMAN-G Plasmid in Yeast
Melissa Gómez [...] Eduardo I. Kessi-Pérez
Jun 20, 2025 1575 Views
A Comparative Protocol for Preserving Deep-Water Marine Invertebrate Tissues: DNA/RNA Shield vs. Liquid Nitrogen for Dual Extraction of High-Quality Nucleic Acids
Ana S. Gomes [...] Olivier Laroche
Nov 20, 2025 1301 Views
Abstract
This protocol presents a modified version of the Filterprep method originally reported in New Biotechnology, adding an optional step to reduce endotoxin levels. Filterprep is a simple, rapid, and cost-effective approach to plasmid DNA purification that couples ethanol precipitation with a single spin-column filtration step, eliminating chaotropic salts and silica binding. The formulations and parameters are fully transparent and do not rely on proprietary buffers, using only standard laboratory reagents and widely available miniprep columns. Under matched conditions, the method recovers high-purity plasmid DNA with yields up to fivefold higher than those obtained with representative commercial midiprep kits. The workflow is readily adoptable in most molecular biology laboratories and, under routine conditions, can be completed in approximately 40 min. The resulting DNA is suitable for molecular cloning, PCR, sequencing, and other downstream biochemical applications. Endotoxin is a lipopolysaccharide (LPS) found in the outer membrane of Gram-negative bacteria and may carry over during plasmid preparation. For experiments requiring lower endotoxin input, an optional modification resuspends the DNA pellet in a Triton X-114 wash buffer before column loading to decrease lipopolysaccharide carryover. The method is modular and extensible, allowing adjustment of precipitation and wash conditions, variation in the number of washes, selection of alternative column formats, and integration of endotoxin-reduction modules without altering the core principle. These features facilitate troubleshooting and quality control, enable scaling from routine batches to larger culture volumes and higher throughput, and allow seamless integration with existing workflows.
Key features
• Modular, chaotrope-free workflow [1] combining ethanol precipitation and single-column cleanup; transparent chemistry allows RNase, endotoxin reduction, or extra washes without changing the core principle.
• Uses only standard reagents, a microcentrifuge, and common miniprep columns, with no proprietary kits, vacuum manifolds, or specialized equipment, enabling broad adoption across laboratories.
• Extensible across scales from small miniprep volumes to larger cultures while remaining compatible with cloning, PCR, sequencing, and transfection-grade applications.
• Optional low-endotoxin modification resuspends the DNA pellet in Triton X-114 wash buffer before column filtration to reduce lipopolysaccharide carryover.
Keywords: Plasmid DNA purificationGraphical overview
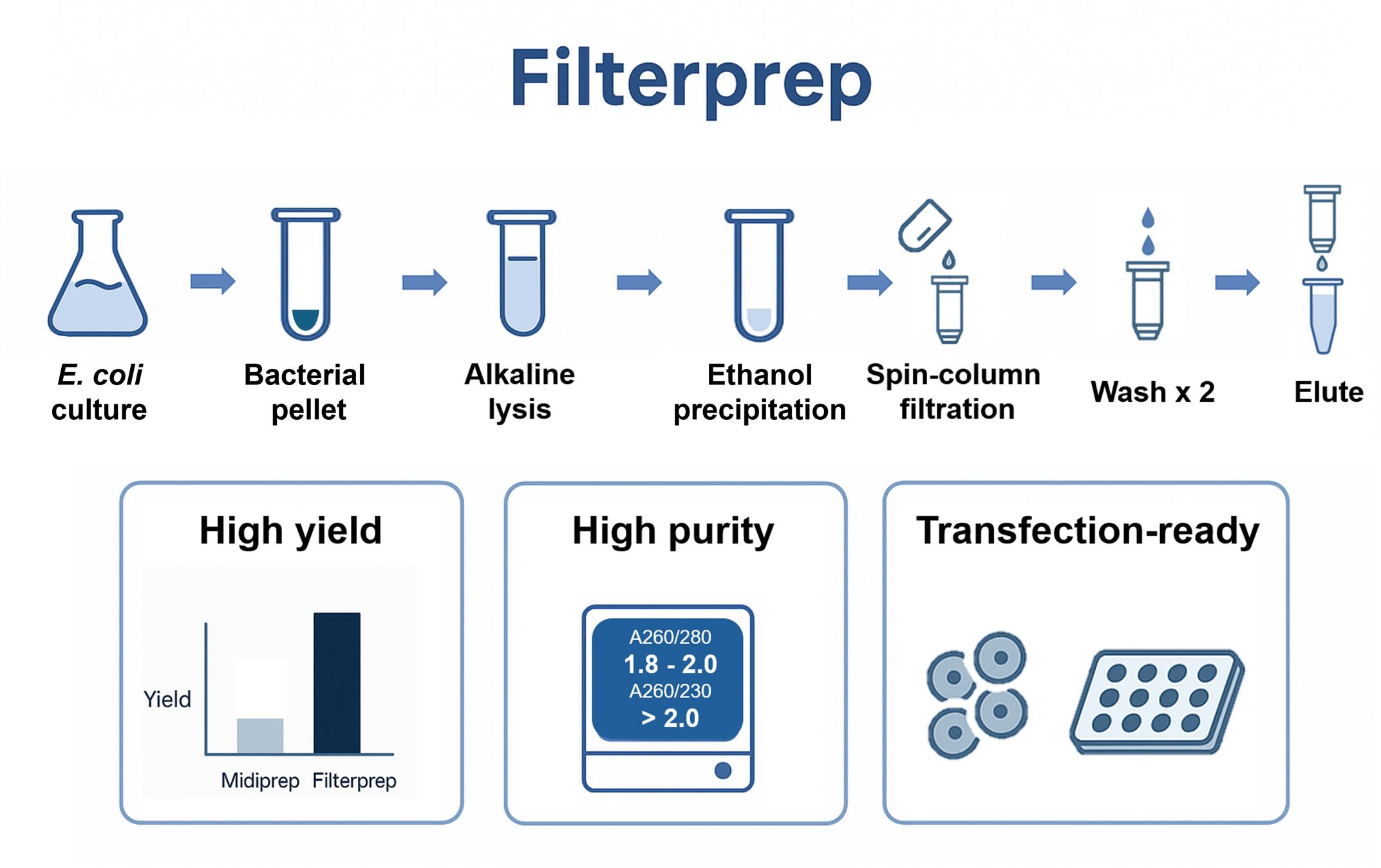
Filterprep hybrid workflow for plasmid purification. Chaotrope-free workflow using ethanol precipitation for capture and a spin column for transfer and washing, delivering high yield and purity with broad downstream compatibility.
Background
Plasmid DNA purification is foundational to molecular cloning, genome engineering, and functional genomics [2–5]. Standard workflows couple alkaline lysis with silica spin columns or magnetic beads. While convenient, these approaches often rely on proprietary formulations and chaotropic salts, which increase cost and limit flexibility [6]. Traditional non-column methods, such as phenol/chloroform extraction with ethanol precipitation or CsCl/ethidium bromide ultracentrifugation, are feasible but require organic solvents and waste handling, involve lengthy and operator-sensitive procedures, and show greater batch-to-batch variability, which reduces suitability for high-throughput, routine use [7,8].
Filterprep combines ethanol precipitation with a single spin column used as a physical filter rather than a silica-binding matrix. The chemistry is transparent and chaotrope-free, relies only on routine reagents and widely available miniprep columns, and is straightforward to implement across laboratories [1]. Under matched conditions, it yields high-purity plasmid DNA and can outperform representative commercial midiprep kits while remaining compatible with molecular cloning, PCR, sequencing, and transfection [9–11].
Endotoxin is a lipopolysaccharide (LPS) derived from the outer membrane of Gram-negative bacteria and may be carried over into plasmid DNA preparations. Although such background levels are generally acceptable for routine applications, some experimental contexts, such as the transfection of sensitive cell lines or preclinical testing, may require reduced endotoxin content. To address this, an optional modification resuspends the DNA pellet in a Triton X-114 wash buffer prior to column loading, thereby decreasing lipopolysaccharide carryover. Prior studies have shown that incorporating a nonionic surfactant such as Triton X-114 directly into the wash buffer can reduce endotoxin carryover without compromising recovery, consistent with the micellar sequestration of LPS [12–14]. This step fits within the existing timeline and requires no additional equipment, making it practical when lower endotoxin input is desired.
Compared with silica-based kits, the protocol reduces dependence on chaotropic salts and proprietary buffers, lowers per-sample cost, and facilitates troubleshooting and quality control, while also avoiding the organic solvents and ultracentrifugation required by traditional non-column methods. These properties shorten hands-on time, reduce batch-to-batch variability, and improve process predictability. Overall, Filterprep supports routine plasmid preparation and method development. Because of its modular design and parameter transparency, it has the potential to extend from routine scale to larger culture volumes and higher throughput by proportionally increasing precipitation and wash volumes, running multiple columns in parallel, or using higher-capacity formats. It can also serve as an upstream pretreatment before downstream polishing and stringent endotoxin-control modules, providing a pathway toward pilot-scale development and industrial application.
Materials and reagents
Biological materials
1. Fast-TransTM competent E. coli DH5α cells (Protech, catalog number: PT-FTDH5)
2. pEGFP-C1 plasmid (Clontech, catalog number: 6084-1)
Reagents
1. LB broth (Miller) (BioShop, catalog number: LBL407)
2. Trizma® base (Sigma, catalog number: T1503)
3. EDTA (BioShop, catalog number: EDT001)
4. Thymolphthalein (Sigma, catalog number: 114553)
5. RNase A (100 mg/mL) (QIAGEN, catalog number: 19101)
6. Hydrochloric acid (HCl) (Merck, catalog number: 1.13134)
7. Potassium acetate (KOAc) (BioShop, catalog number: POA301)
8. Glacial acetic acid (Sigma, catalog number: A6283)
9. Sodium dodecyl sulfate (SDS) (Sigma, catalog number: 75746)
10. Sodium hydroxide (NaOH) (Sigma, catalog number: 221465)
11. 95% ethanol (Merck, catalog number: 1.00983)
12. 100% ethanol (Sigma, catalog number: 459844)
13. Triton X-114 (Sigma, catalog number: 93422)
Solutions
1. 1 M thymolphthalein (see Recipes)
2. Solution 1 (see Recipes)
3. Solution 2 (see Recipes)
4. Solution 3 (see Recipes)
5. Wash buffer (see Recipes)
6. Endotoxin removal buffer (ETR Buffer) (see Recipes)
Recipes
1. 1 M thymolphthalein
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Thymolphthalein | 1 M | 4.3 g |
| 100% ethanol | to a final volume of 10 mL |
2. Solution 1
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Tris-HCl, pH 8.0 | 50 mM | 25 mL |
| 0.5 M EDTA, pH 8.0 | 10 mM | 10 mL |
| 1 M thymolphthalein (optional) | 1 mM | 0.5 mL |
| ddH2O | to a final volume of 500 mL |
3. Solution 2
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| NaOH | 0.2 N | 4 g |
| SDS | 1% | 5 g |
| ddH2O | to a final volume of 500 mL |
3. Solution 3
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| KOAc | 3 M | 147.2 g |
| ddH2O | 400 mL | |
| Glacial acetic acid | Titrate to pH 5.5 | |
| ddH2O | to a final volume of 500 mL |
4. Wash buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Tris-HCl, pH 7.5 | 10 mM | 5 mL |
| 100% ethanol | 80% | 400 mL |
| ddH2O | n/a | 95 mL |
| Total | n/a | 500 mL |
5. Endotoxin removal buffer (ETR buffer)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Wash buffer | n/a | 99.8 mL |
| Triton X-114 | 0.2% | 0.2 mL |
Laboratory supplies
1. 14 mL culture tube (Greiner Bio-One, catalog number: 187261)
2. 250 mL baffled flask (DURAN, catalog number: 212833655)
3. KimwipesTM (Kimberly-Clark, catalog number: 34155)
4. 20 mL syringe (TERUMO, catalog number: SS-20L)
5. 50 mL conical centrifuge tube (Greiner Bio-One, catalog number: 227261)
6. PrestoTM Mini Plasmid kit (Geneaid, catalog number: PDH300) (only spin columns were used; buffers were not included in this protocol)
Equipment
1. PH meter (METTLER TOLEDO, model: F20)
2. Centrifuge (Eppendorf, model: 5427R; rotor FA-45-30-11)
3. Centrifuge (KUBOTA, model: 5922; rotor RA-410M3)
4. NanoDrop (Thermo Fisher Scientific, model: NanoDrop One)
Procedure
A. Preparation of transformed bacteria
1. This protocol was tested using E. coli DH5α carrying the high-copy plasmid pEGFP-C1, a commonly used mammalian expression vector. While compatible with other vectors and strains, yield and lysis behavior may vary depending on plasmid size, copy number, and host background.
Note: Plasmids larger than 10 kb or low-copy constructs may require modified culture time or lysis buffer volumes (see General note 2).
2. Transform E. coli DH5α with pEGFP-C1 using the standard heat-shock method for chemically competent cells as described in [15]. Briefly, mix 50–100 ng of plasmid DNA with 100 μL of competent cells, incubate on ice for 30 min, heat-shock at 42 °C for 45 s, return to ice for 2 min, recover in 400 μL of LB at 37 °C for 1 h, and plate on LB agar with the appropriate antibiotic. Incubate overnight at 37 °C.
3. Pick a single, well-isolated colony and inoculate 3 mL of LB broth with the corresponding antibiotic in a 14 mL culture tube. Incubate at 37 °C with shaking (200–250 rpm) for 6–8 h.
4. Inoculate 50 mL of fresh LB broth (with antibiotic) in a 250 mL baffled flask with 0.5 mL of the pre-culture. Incubate overnight (12–16 h) at 37 °C with shaking at 200–250 rpm until saturation (OD600 ≈ 2.0–3.0).
Critical: Use a flask at least 3–5 times the culture volume to ensure proper aeration.
B. Cell harvest and resuspension
1. Transfer 50 mL of the overnight bacterial culture to a 50 mL conical tube.
2. Centrifuge at 6,000× g for 10 min at 4 °C in a refrigerated centrifuge. Carefully remove the supernatant without disturbing the pellet.
3. Prepare solution 1 containing RNase A immediately before use by adding 4 µL of RNase A (100 mg/mL) to 4 mL of solution 1 (see Recipes) and mixing by gentle inversion. Resuspend the pellet completely in 4 mL of solution 1 with RNase A until no visible clumps remain (Figure 1A).
Note: Buffer volumes can be adjusted according to culture size (approximately 1:20 ratio) (see General note 3).
Critical: Ensure complete resuspension to enable efficient lysis in the next step.
See Troubleshooting, problem 1.
C. Alkaline lysis and neutralization
1. Add 4 mL of solution 2 (see Recipes). Immediately mix by gently inverting the tube 4–6 times to homogenize. Incubate at room temperature for 3 min. Do not exceed 5 min (Figure 1B).
Caution: Solution 2 is alkaline and contains SDS. Wear gloves and eye protection.
Critical: Do not vortex; vigorous mixing shears genomic DNA and lowers plasmid purity.
Note: If thymolphthalein indicator is included, the suspension turns blue upon lysis (see General note 4).
See Troubleshooting, problem 1.
2. Add 4 mL of solution 3 (see Recipes). Immediately mix by gently inverting the tube 4–6 times to ensure thorough neutralization. Incubate at room temperature for 10 min (Figure 1C).
Note: If thymolphthalein indicator is included, mix until the blue color disappears completely, indicating full neutralization.
Critical: Incomplete neutralization reduces clarification and yield.
See Troubleshooting, problem 2.
3. Centrifuge at ≥13,200× g for 10 min at 4 °C to pellet cell debris and precipitated proteins. Confirm that the supernatant is clear; if cloudy, repeat the centrifugation.
Critical: A turbid supernatant indicates insufficient clarification.
See Troubleshooting, problem 2.
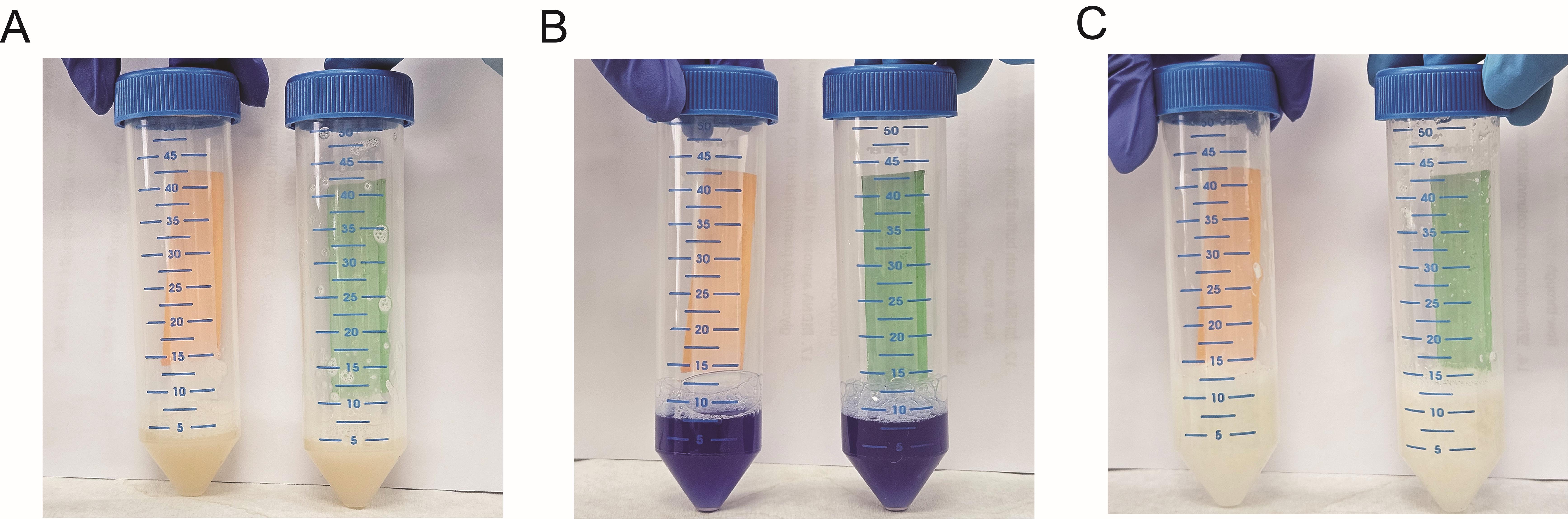
Figure 1. Color transitions during the resuspension-lysis-neutralization workflow. (A) Pellet after thorough resuspension in solution 1. (B) Lysate color after thorough mixing with solution 2. (C) Lysate color after thorough mixing with solution 3.
D. Syringe filtration and ethanol precipitation
1. Transfer the clear supernatant to a 20 mL syringe preloaded with a single sheet of KimwipesTM (folded to fit) as a filter (Figure 2A–C). Gently insert the plunger and collect the filtrate into a 50 mL tube (Figure 2D). This step helps remove residual particulate matter or diffuse macromolecular aggregates that may interfere with column flow or compromise plasmid purity.
Critical: Apply steady, gentle pressure to avoid channeling (see General note 5).
Note: Place the syringe outlet close to the tube wall to minimize aerosol formation.
See Troubleshooting, problem 3.
2. Precipitate DNA by adding 1 volume (typically ~12 mL) of 95% ethanol equal to the filtrate volume. Mix thoroughly by inversion and centrifuge at ≥13,200× g for 10 min at 4 °C. Carefully decant the supernatant.
Pause point: The DNA pellet may remain submerged in 95% ethanol at 4 °C for up to 1 h or at -20 °C for up to 24 h before proceeding.
Note: For diluted or low-copy samples, increase ethanol to 1.2–1.5× or briefly chill before spinning; up to 2.0× may be used with additional washing/dry-spin (see General note 6).
See Troubleshooting, problem 4.
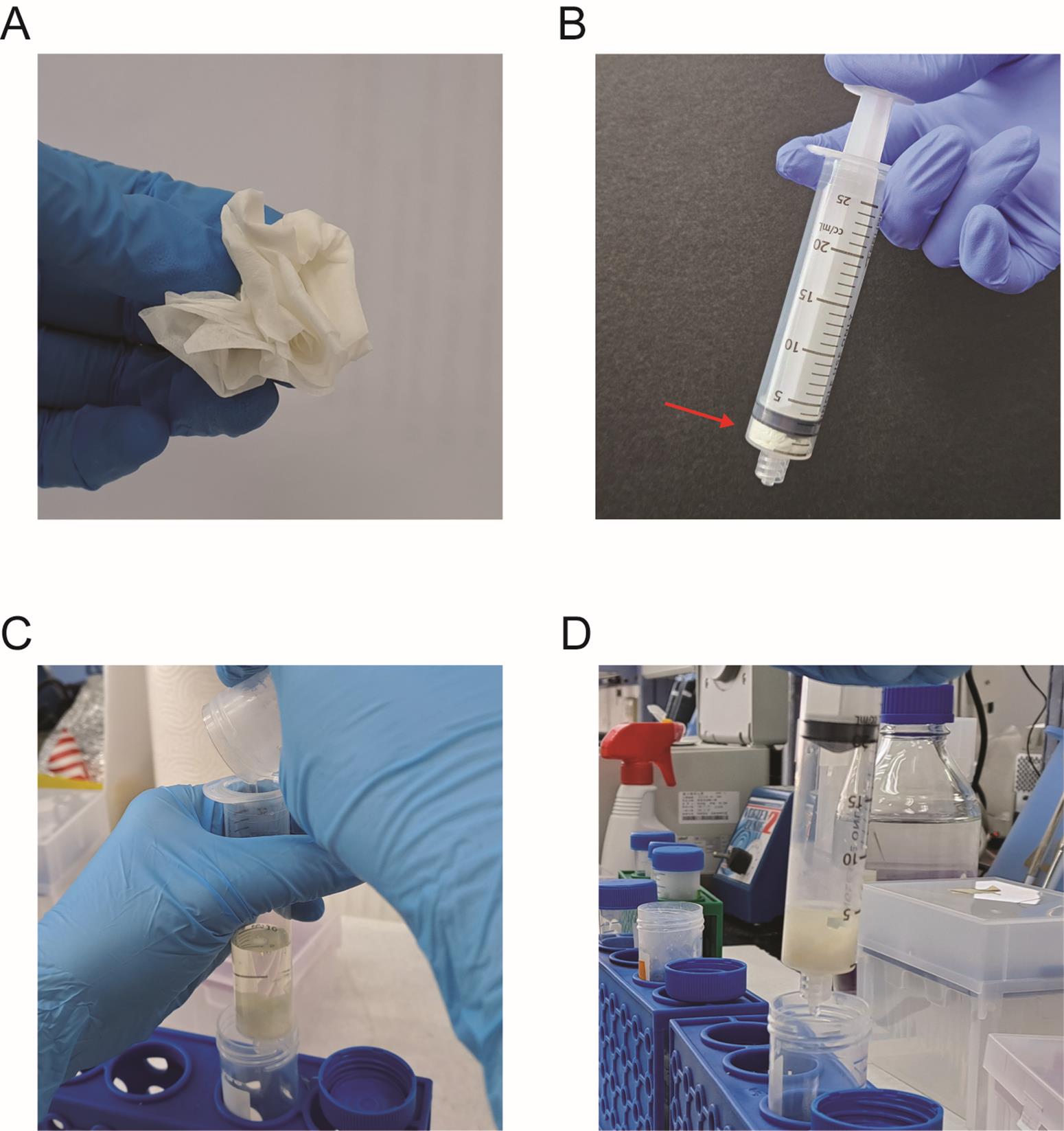
Figure 2. Schematic of supernatant filtration using KimwipesTM. (A) Folding of a KimwipesTM tissue. (B) A 20 mL syringe packed with the folded KimwipesTM tissue (indicated by a red arrow). (C) Pour the clarified supernatant into the packed 20 mL syringe. (D) Insert the plunger and press gently to filter into a 50 mL tube.
E. Column capture and washing
1. Add 750 μL of wash Buffer to the DNA pellet and gently resuspend by pipetting until fully dispersed. Transfer the entire suspension to a miniprep spin column (PrestoTM Mini Plasmid Kit) placed in its collection tube. Equivalent standard silica columns from other commercial sources can also be used without affecting yield or purity.
Note: Slightly trimming the pipette tip can prevent clogging by the pellet. If endotoxin reduction is desired, you may use ETR buffer instead of wash buffer in this loading step (see General note 7). If the sample volume exceeds the column capacity, the same column may be reloaded after the first centrifugation step, provided the flowthrough is discarded between loads. Also, see General note 8 regarding splitting large loads across two columns.
See Troubleshooting, problem 5.
2. Centrifuge the column at ≥13,200× g for 30 s at room temperature (20–25 °C). Discard the flowthrough.
3. Add 750 µL of wash buffer to the column. Centrifuge at ≥13,200× g for 30 s at room temperature (20–25 °C). Discard the flowthrough.
4. Repeat step E3 once more.
5. Centrifuge at ≥13,200× g for 1 min at room temperature (20–25 °C) to remove residual wash buffer.
Critical: This dry spin removes residual ethanol that can inhibit downstream enzymes.
F. Elution and storage
1. Place the spin column in a clean 1.5 mL microcentrifuge tube. Add 200 µL of nuclease-free water to the membrane, let stand for 2 min at room temperature, then centrifuge at ≥13,200× g for 2 min at room temperature (20–25 °C) to elute DNA.
Note: Prewarming the eluent to 50–60 °C and a 2–5 min on-membrane incubation can improve yield (see General note 9). A second elution with an additional 100–200 µL increases total mass at the cost of lower concentration.
See Troubleshooting, problem 6.
2. Measure DNA concentration and purity using a spectrophotometer. Record A260/A280 and A260/A230 values. Store DNA at -20 °C (see General note 10).
See Troubleshooting, problem 7.
Validation of protocol
This protocol (or parts of it) has been used and validated in the following research article:
Cheng et al. [1] The Filterprep: A simple and efficient approach for high-yield, high-purity plasmid DNA purification. New Biotechnology.
In addition to the validation reported in our New Biotechnology publication [1], previous studies have shown that Triton X-114 effectively removes lipopolysaccharide (LPS) contaminants from biological preparations [12–14]. Although no quantitative endotoxin assay (e.g., LAL test) was performed in this protocol, spectrophotometric measurements (OD 260/280 and OD 260/230 ratios) indicated that inclusion of Triton X-114 did not noticeably affect plasmid yield or purity (Table 1). This optional wash step further demonstrates the modular nature of the Filterprep workflow, allowing users to incorporate additional modules, such as endotoxin reduction, while maintaining its core principles of precipitation and filtration.
Table 1. Comparison of plasmid DNA purification methods using Qiagen kits and Filterprep protocols with or without the Triton X-114 wash step
| Properties | Midiprep (QIAGEN) | Filterprep (without Triton X-114 wash) | Filterprep (with Triton X-114 wash) |
|---|---|---|---|
| Bacteria culture (mL) | 50 | 50 | 50 |
| Average yield (μg) | 176.7 ± 21.75 | 1,432 ± 281.1 | 1,381 ± 132.9 |
| OD 260/280 | 1.86 | 1.97 | 1.95 |
| OD 260/230 | 2.62 | 2.47 | 2.45 |
| Time (min) | 150 | <40 | <40 |
| Costs per μg DNA (cents) | 9.8 | 0.1 | 0.1 |
General notes and troubleshooting
General notes
1. Principle and positioning: This method recovers plasmid DNA by ethanol precipitation, then uses a miniprep column primarily as a support for transfer and washing, not for chaotropic salt–driven silica binding. No high-chaotrope binding step is required; residual salts mainly derive from the neutralized lysate.
2. Culture conditions and copy number: There is no hard upper limit on culture volume. Yield depends on plasmid copy number, host genotype (use endA− strains such as DH5α or TOP10), and maintenance of antibiotic selection. When the OD plasmid copy is low, increase the culture volume or run multiple flasks in parallel and combine them before processing. Scale all buffer volumes proportionally while keeping the same lysis/neutralization times and g-forces (see General Note 3). When the supernatant volume becomes large, perform syringe filtration and ethanol precipitation in batches. Split the resuspended pellet across one or more columns to maintain flow and wash efficiency (see General Note 7). Observe tube and rotor rated capacities and g-limits.
3. Buffer ratios and linear scaling: Buffer volumes are scaled relative to the culture volume at an approximate ratio of 1:20. This ratio follows the standard alkaline lysis principle established by Birnboim and Doly [2], ensuring complete lysis and consistent plasmid recovery. For a 50 mL bacterial culture, use 4 mL each of solution 1 (resuspension), solution 2 (lysis), and solution 3 (neutralization). Add RNase A to solution 1 immediately before use to reach a final concentration of 100 µg/mL [e.g., 4 µL of RNase A (100 mg/mL) per 4 mL of solution 1]. For smaller or larger input volumes, adjust all buffer and RNase A volumes proportionally and maintain consistent mixing, incubation, and centrifugation conditions to ensure comparable plasmid yield and purity.
4. The indicator is auxiliary: With thymolphthalein, the lysate turns blue upon alkaline lysis and becomes colorless after neutralization. Color depends on temperature and ionic strength, so rely on timing and thorough mixing; use color only as an aid.
5. Syringe–Kimwipe filtration: Fold a single Kimwipes™ sheet to line the syringe wall. Filter the sample directly. If channeling occurs, optionally pre-wet the pad with a small amount of supernatant before full filtration.
6. Ethanol precipitation parameters: Default is 1.0× volume of 95% ethanol. To enhance recovery, first increase ethanol to 1.2–1.5× or chill briefly at 4 °C or -20 °C before spinning. Reserve up to 2.0× for very dilute or low-copy samples and, if used, add one extra 750 µL wash buffer rinse and extend the dry spin to 1–2 min to minimize salt and ethanol carryover.
7. Endotoxin reduction option: When high transfection efficiency is required, substitute the ETR buffer for wash buffer at the pellet-to-column transfer step. Expect lower endotoxin with a modest decrease in total yield.
8. Role of the column: Disperse the DNA pellet in 750 µL of wash buffer before loading. If the suspension is large, split across two columns to avoid slow flow.
9. Elution and storage: Incubating nuclease-free water at 50–60 °C on the membrane for 2–5 min improves yield. A second elution increases total mass but reduces concentration. For long-term storage, elute with TE pH 8.0. Blank spectrophotometer with the same eluent.
10. Quality metrics and downstream compatibility: Typical A260/A280 is 1.8–2.0, and A260/A230 is above 2.0. DNA is suitable for restriction digest, PCR, sequencing, and most transfections. If transfection is poor, check for residual ethanol or endotoxin first.
Troubleshooting
Problem 1: Lysate becomes highly viscous, and final DNA purity is low.
Possible cause: Overmixing or over-incubation during alkaline lysis causes genomic DNA shearing and co-purification.
Solution: Limit lysis to 3 min at room temperature. Mix by gentle inversion only (4–6 times). Do not vortex. Proceed promptly to neutralization.
Problem 2: Supernatant remains cloudy, or blue does not fully disappear after neutralization.
Possible cause: Incomplete mixing with solution 3 or insufficient clarification spin.
Solution: Invert thoroughly immediately after adding solution 3 until homogenized; with the indicator, mix until completely colorless. Centrifuge at ≥13,200× g for 10–15 min at 4 °C; repeat if still turbid.
Problem 3: Weak or invisible DNA pellet after ethanol precipitation.
Possible cause: Ethanol proportion too low or temperature too warm for efficient nucleation; sample is very dilute or low copy.
Solution: Increase ethanol from 1.0× to 1.2–1.5× (95%–100% ethanol) or chill the mixture at 4 °C or -20 °C for 10–20 min before centrifugation. If still insufficient, increase to a maximum of 2.0× and add an extra 750 µL wash buffer rinse with an extended 1–2 min dry spin to limit salt and ethanol carryover (see General Note 6).
Problem 4: Syringe–Kimwipe filtration is very slow or shows channeling.
Possible cause: Filter folded too thick, uneven packing, or carried-over flocculent material.
Solution: Refold a single sheet to fit snugly, pre-wet, and apply steady pressure. Replace the filter or perform a brief additional clarification spin before filtration if needed.
Problem 5: Column flow is slow or the column clogs.
Possible cause: Pellet not fully dispersed, or load exceeds practical capacity.
Solution: Fully disperse in 750 µL of wash buffer before loading; trim the pipette tip to ease transfer. Split across two columns if needed.
Problem 6: Low elution yield or concentration.
Possible cause: Insufficient on-membrane incubation or residual ethanol on the membrane.
Solution: Elute with prewarmed nuclease-free water (50–60 °C) and incubate for 2–5 min on the membrane. Consider a second elution.
Problem 7: Good yield but low transfection efficiency.
Possible cause: Residual ethanol or endotoxin.
Solution: Extend the dry spin to 2 min. For sensitive applications, use ETR buffer at the pellet-to-column step.
Acknowledgments
We thank colleagues for helpful discussions and technical assistance. Author contributions: Conceptualization, C.C.; Investigation, Y.L., Y.S.; Writing—Original Draft, C.C.; Writing—Review & Editing, C.C.; Funding acquisition, C.C.; Supervision, C.C. This work was supported by the National Science and Technology Council, Taiwan (NSTC 114-2311-B-A49-001-) and the Yen Tjing Ling Medical Foundation (CI-114-19). This protocol was described and validated in our original research article in New Biotechnology (2025, DOI: 10.1016/j.nbt.2025.09.003), with an added optional endotoxin-reduction step.
Competing interests
The authors declare no conflicts of interest.
References
- Cheng, Y. H., Wu, Y. J., Shih, Y. C., Lin, Y. Q., Chang, Y. W., Wu, Y. H., Hu, E. W., Ku, R. H., Wu, C. M., Wu, S. C., et al. (2025). The Filterprep: A simple and efficient approach for high-yield, high-purity plasmid DNA purification. New Biotechnol. 90: 77–87. https://doi.org/10.1016/j.nbt.2025.09.003
- Birnboim, H. and Doly, J. (1979). A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7(6): 1513–1523. https://doi.org/10.1093/nar/7.6.1513
- Boom, R., Sol, C. J., Salimans, M. M., Jansen, C. L., Wertheim-van Dillen, P. M. and van der Noordaa, J. (1990). Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 28(3): 495–503. https://doi.org/10.1128/jcm.28.3.495-503.1990
- Clewell, D. B. and Helinski, D. R. (1969). Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci USA. 62(4): 1159–1166. https://doi.org/10.1073/pnas.62.4.1159
- Colman, A., Byers, M. J., Primrose, S. B. and Lyons, A. (1978). Rapid Purification of Plasmid DNAs by Hydroxyapatite Chromatography. Eur J Biochem. 91(1): 303–310. https://doi.org/10.1111/j.1432-1033.1978.tb20966.x
- Marko, M., Chipperfield, R. and Birnboim, H. (1982). A procedure for the large-scale isolation of highly purified plasmid DNA using alkaline extraction and binding to glass powder. Anal Biochem. 121(2): 382–387. https://doi.org/10.1016/0003-2697(82)90497-3
- Green, M. R. and Sambrook, J. (2017). Precipitation of DNA with Isopropanol. Cold Spring Harb Protoc. 2017(8): pdb.prot093385. https://doi.org/10.1101/pdb.prot093385
- He, M., Kaderbhai, M. A., Adcock, I. and Austen, B. M. (1991). An improved and rapid procedure for isolating RNA-Free Escherichia coli plasmid DNA. Genet Anal Tech Appl. 8(3): 107–110. https://doi.org/10.1016/1050-3862(91)90045-s
- Chen, T. W., Liao, H. W., Noble, M., Siao, J. Y., Cheng, Y. H., Chiang, W. C., Lo, Y. T. and Chang, C. T. (2024). Human DCP1 is crucial for mRNA decapping and possesses paralog-specific gene regulating functions. eLife. 13: e94811. https://doi.org/10.7554/elife.94811
- Cheng, Y. H., Chen, T. W., Chiang, W. C., Kuo, J. C., Ho, Y. S., Noble, M. and Chang, C. T. (2025). EDC4 C-terminal domain scaffolds P-body assembly and links P-body dynamics to p53-mediated tumor suppression. RNA. 31(8): 1176–1194. https://doi.org/10.1261/rna.080561.125
- Weber, R. and Chang, C. T. (2024). Human DDX6 regulates translation and decay of inefficiently translated mRNAs. eLife. 13: e3. https://doi.org/10.7554/elife.92426.3
- Kiesewetter, A., Gupta, A., Heinen‐Kreuzig, A., Greenhalgh, T. and Stein, A. (2023). Improved endotoxin removal using ecofriendly detergents for intensified plasmid capture. Biotechnol Progr. 39(6): e3375. https://doi.org/10.1002/btpr.3375
- Koziel, D., Michaelis, U. and Kruse, T. (2018). Broad application and optimization of a single wash-step for integrated endotoxin depletion during protein purification. J Chromatogr B. 1091: 101–107. https://doi.org/10.1016/j.jchromb.2018.05.029
- Petsch, D. (2000). Endotoxin removal from protein solutions. J Biotechnol. 76: 97–119. https://doi.org/10.1016/s0168-1656(99)00185-6
- Green, M. R., Sambrook, J. and Sambrook, J. (2012). Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, N.Y. ISBN: 9781936113415.
Article Information
Publication history
Received: Oct 9, 2025
Accepted: Nov 18, 2025
Available online: Nov 26, 2025
Published: Dec 20, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Lin, Y., Shih, Y. and Chang, C. (2025). Plasmid DNA Purification Using Filterprep With an Optional Endotoxin Removal Step. Bio-protocol 15(24): e5547. DOI: 10.21769/BioProtoc.5547.
Category
Molecular Biology > DNA > DNA extraction
Microbiology > Microbial genetics > Plasmid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









