- EN - English
- CN - 中文
Orthogonal Temperature-Related Intensity Change and Time-Resolved Förster Resonance Energy Transfer High-Throughput Screening Platform for the Discovery of SLIT2 Binders
发布: 2026年02月20日第16卷第4期 DOI: 10.21769/BioProtoc.5604 浏览次数: 73
评审: Anonymous reviewer(s)

相关实验方案
基于蛋白筛选策略从合成文库中分离抗原特异性纳米抗体:结合 MACS 的酵母展示筛选与 FLI-TRAP 方法
Apisitt Thaiprayoon [...] Dujduan Waraho-Zhmayev
2026年01月20日 433 阅读
Abstract
SLIT2 is a secreted glycoprotein implicated in axon guidance, immune modulation, and tumor biology, whose extracellular and glycosylated nature can complicate conventional biophysical screening workflows. Here, we provide a complete, step-by-step protocol for an orthogonal high-throughput discovery pipeline that integrates temperature-related intensity change (TRIC) as a solution-based primary binding screen with time-resolved Förster resonance energy transfer (TR-FRET, homogeneous time-resolved fluorescence format) as a functional assay for inhibition of the SLIT2–ROBO1 interaction. The workflow is designed to be fast and convenient, uses low reaction volumes and low nanomolar protein concentrations to minimize material use, and includes built-in quality control steps to support reproducible hit triage. In TRIC (NanoTemper Dianthus), binding is detected as temperature-dependent fluorescence intensity changes of a labeled target protein under an infrared (IR)-mediated thermal gradient, enabling immobilization-free detection of small-molecule interactions and instrument-assisted filtering of autofluorescent, quenching, or aggregating compounds. Candidate binders are advanced to multi-point TRIC/microscale thermophoresis (MST) measurements on Monolith X to determine binding affinity (Kd). In TR-FRET, disruption of SLIT2–ROBO1 association is quantified by changes in the ratiometric 665/620 nm emission readout, measured with a time delay to suppress short-lived background fluorescence, enabling concentration-response analysis and reporting of relative IC50 values (including partial inhibition behavior where applicable). Although presented using the SLIT2–ROBO1 extracellular interaction as a representative model system, this orthogonal screening strategy is designed to be adaptable to other extracellular protein-protein interactions where minimizing immobilization artifacts and fluorescence interference is critical.
Key features
• End-to-end pipeline: TRIC primary screen → artifact filtering → MST/Monolith X affinity → TR-FRET inhibition validation.
• Homogeneous, immobilization-free assays compatible with 384-well HTS formats.
• Reproducible analysis: standardized thresholds for TRIC hits and relative IC50 reporting for partial inhibitors.
• Generalizable to other extracellular glycoproteins and ligandable protein–protein interactions (PPIs).
Keywords: SLIT2Graphical overview
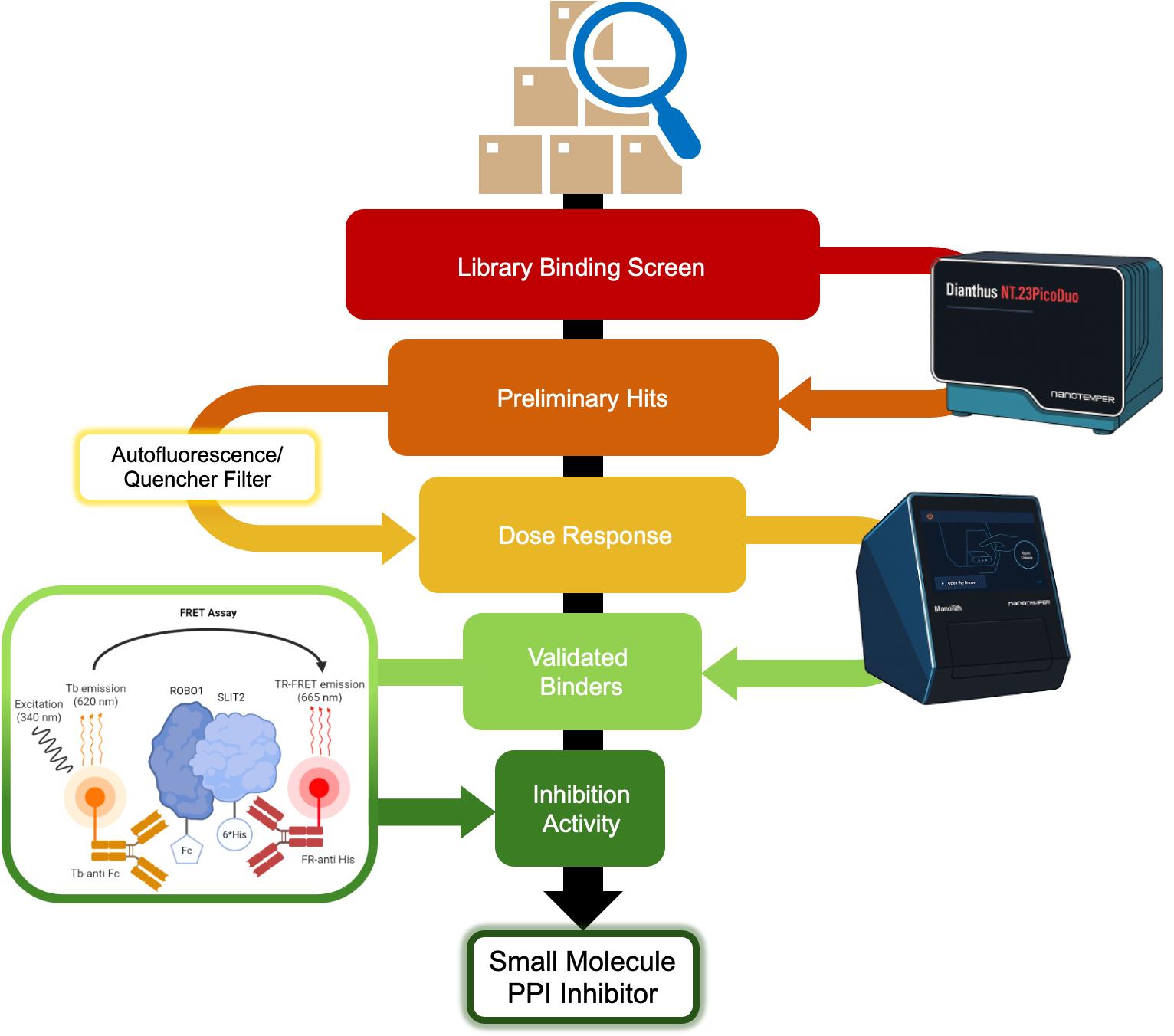
Orthogonal screening workflow for identifying SLIT2 binders and SLIT2–ROBO1 interaction inhibitors. A small molecule library is first evaluated in a single concentration binding screen against fluorescently labeled SLIT2 His using NanoTemper Dianthus (TRIC) in a 384-well format to generate preliminary binding signals. Compounds are then subjected to an artifact filter to remove wells affected by autofluorescence, quenching, or other assay-interfering behavior, yielding a curated set of preliminary hits. These candidates are progressed to dose response binding measurements on NanoTemper Monolith X (TRIC/MST) to confirm concentration-dependent binding and quantify affinity (reported as Kd), defining validated binders. Confirmed binders are then tested in an orthogonal time-resolved Förster resonance energy transfer (TR-FRET) inhibition assay for the SLIT2–ROBO1 extracellular interaction, enabling classification of compounds by inhibition activity and prioritization of candidates with both binding and functional disruption consistent with small-molecule protein–protein interaction (PPI) inhibitor behavior.
Background
The SLIT–ROBO pathway is a conserved signaling axis that regulates cytoskeletal remodeling, axon guidance, and epithelial cell migration [1–4]. Aberrant activation of this pathway has been implicated in cancer progression, angiogenesis, and immune modulation [5–8]. Despite its therapeutic relevance, no small-molecule modulators targeting SLIT2 have been reported until recently, largely due to the extracellular and glycosylated nature of SLIT2 that complicates biophysical screening [7].
To overcome these limitations, we established an orthogonal, immobilization-free screening platform combining temperature-related intensity change (TRIC) for primary binder discovery and time-resolved Förster resonance energy transfer (TR-FRET) for functional inhibition validation of the SLIT2–ROBO1 interaction. In this pipeline, the TRIC screen is followed by concentration-response affinity determination (Kd) for selected hits and TR-FRET concentration-response testing to report relative IC50 values [9,12–14].
This combination leverages two complementary principles: First, TRIC (NanoTemper Dianthus) measures micro-environmental fluorescence changes induced by molecular binding under infrared (IR)-mediated thermal gradients. Because it requires no immobilization and is compatible with crude or glycosylated proteins, TRIC is well-suited for detecting weak or transient interactions typical of extracellular PPIs [11–14]. Second, TR-FRET [homogeneous time-resolved fluorescence (HTRF) format] detects the disruption of a specific protein–protein interaction using donor-acceptor energy transfer between terbium (Tb3+) and d2 fluorophores. A time delay (integration delay) is applied after excitation so that short-lived background signals decay before detection, while the long-lived terbium emission is retained. The assay is read ratiometrically as a 665/620 nm emission ratio, which improves robustness to well-to-well intensity variation and helps reduce sensitivity to nonspecific intensity fluctuations [9].
Dimethyl sulfoxide (DMSO) content is an important practical variable in both thermophoretic and fluorescence-based assays. In this workflow, DMSO was evaluated across a 0%–10% range, and intermediate levels (approximately 2%–5%) provided the most consistent signal-to-noise under the conditions described here. Lower than 2% or higher than 5% DMSO introduced increased noise in our hands (data not shown). For screening, we therefore fix the in-well DMSO at 2% across all wells and controls to minimize viscosity and matrix-driven variability.
Proteins were sourced in formats compatible with both TRIC labeling and TR-FRET detection. SLIT2 is used as a His-tagged recombinant protein to enable selective TRIC labeling using an NTA-based dye (RED-tris-NTA 2nd Gen) and recognition by the anti-6His d2 acceptor antibody in the TR-FRET assay. ROBO1 is used as an Fc-tagged recombinant protein so that the complex can be detected by the anti-human IgG-Tb donor antibody. A specific consideration of the ROBO1-Fc format is Fc-mediated dimerization, which can increase apparent avidity; however, under the low nanomolar protein concentrations used in this protocol, we did not observe obvious assay artifacts attributable to Fc valency. Reported potencies should be interpreted in the context of the constructs and assay geometry used here. If a monomeric ROBO1 construct is available, it can be substituted, but the TR-FRET signal window and protein ratios should be re-optimized because tag position and spacing influence FRET efficiency.
The workflow (as shown in the Graphical overview) proceeds as follows:
• Step 1: TRIC single-point screening of a focused small molecule library (TargetMol Lipid Metabolism Library) against fluorescently labeled SLIT2 in a 384-well format using NanoTemper Dianthus (Figure 1).
• Step 2: Removal of assay-interfering compounds by filtering wells flagged for autofluorescence, quenching, or aggregation using instrument quality control outputs, yielding preliminary hits.
• Step 3: Confirmation of binding and determination of binding affinity (Kd) by multi-point TRIC/microscale thermophoresis (MST) dose response measurements using NanoTemper Monolith X.
• Step 4: Orthogonal TR-FRET assay to evaluate the effect of confirmed binders on SLIT2–ROBO1 association, generating concentration response curves and reporting relative IC50 values (Figure 2).
This platform enables quantitative and reproducible triage of early hits while minimizing false positives due to fluorescence artifacts, aggregation, or thermal instability. As a representative example, we previously identified bexarotene as a reproducible SLIT2 binder (Kd = 2.62 μM) that partially inhibits SLIT2–ROBO1 binding (IC50 ≈ 77 μM). This example underlies the optimization steps detailed below and serves as a benchmark for troubleshooting and expected performance metrics [1,2].
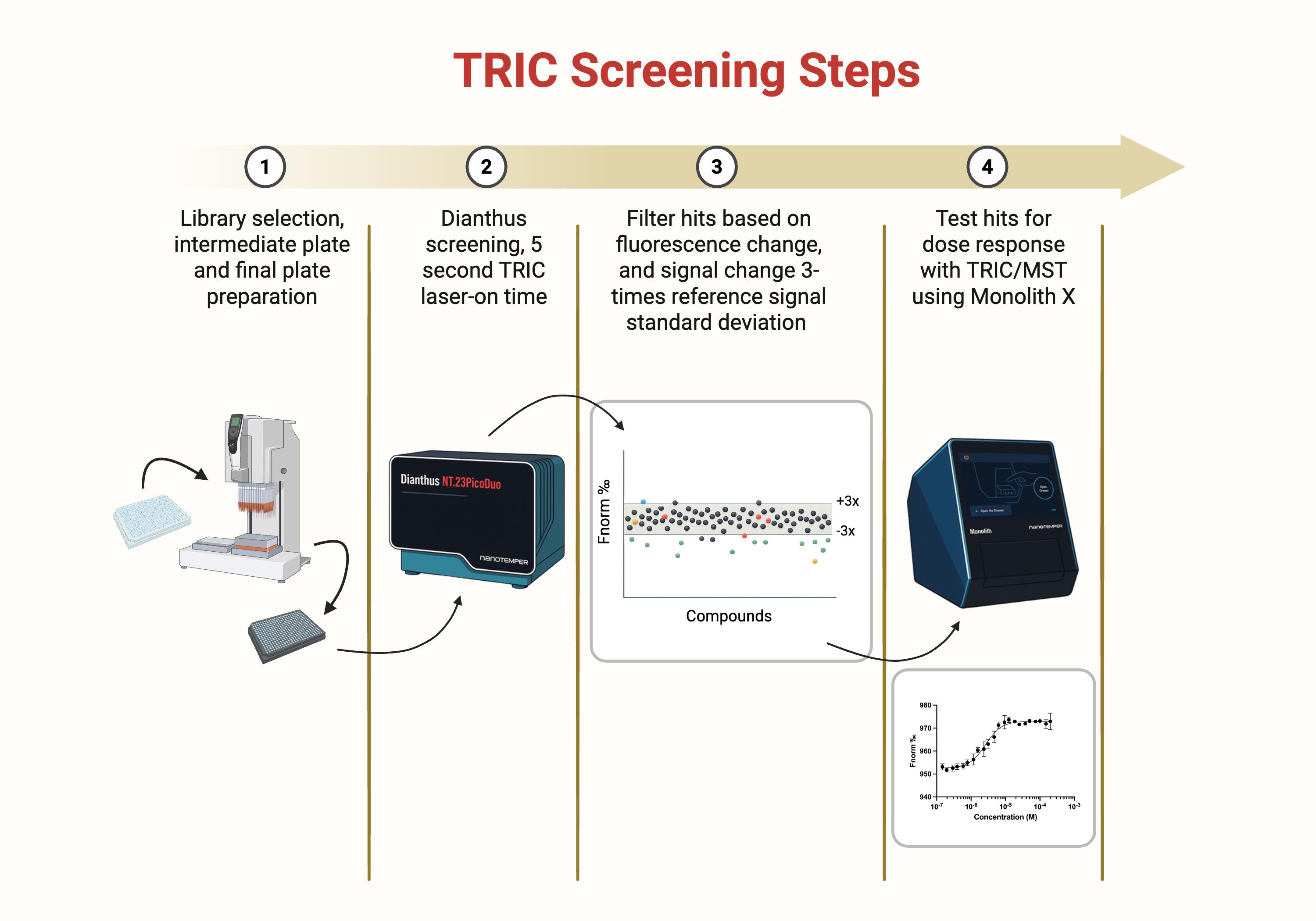
Figure 1. Temperature-related intensity change (TRIC) screening steps overview. Schematic overview of the four-step screening pipeline used to identify SLIT2 binders by TRIC and confirm binding by dose response. (1) Compound library selection and preparation of intermediate dilution plates and final assay plates. (2) Primary single-point TRIC screen on the Dianthus platform using a 5 s laser-on time per well to measure changes in fluorescence signal (Fnorm) relative to vehicle controls. (3) Hit filtering based on fluorescence behavior and magnitude of signal change, retaining compounds that exceed a predefined threshold (for example, ±3× the standard deviation of the reference control distribution) while removing wells flagged for autofluorescence, quenching, or aggregation. (4) Concentration response testing of shortlisted hits using TRIC and microscale thermophoresis (MST) on the Monolith X to generate binding curves and quantify affinity from fitted dose response data.
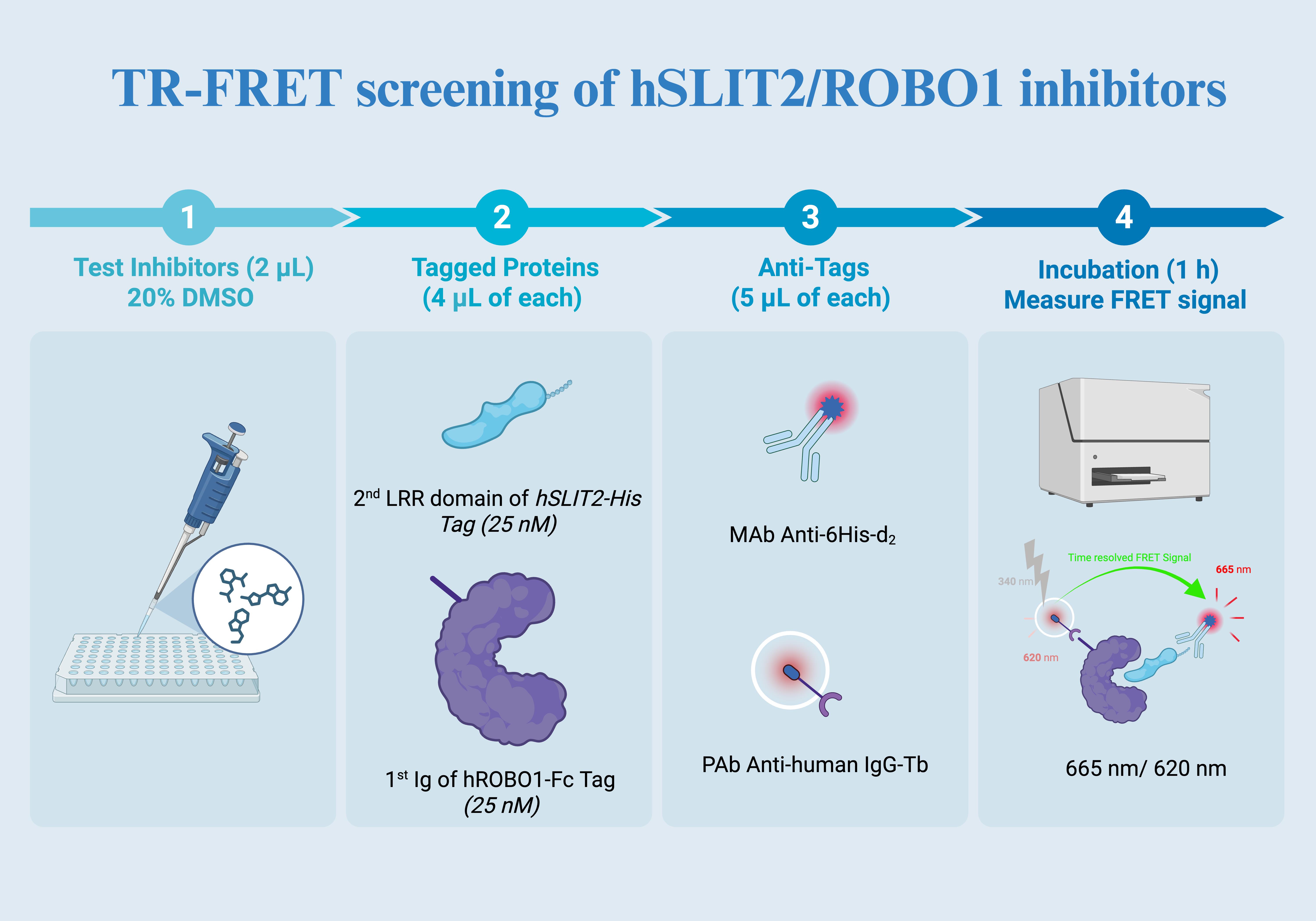
Figure 2. TR-FRET screening overview. (1) Test compounds are dispensed into assay wells (2 μL; 20% DMSO stock) to achieve the desired final compound concentration and DMSO content after all additions. (2) Tagged proteins are added (4 μL of each) to form the complex, using the second LRR domain of hSLIT2 carrying a His tag (25 nM) and the first Ig domain of hROBO1 carrying an Fc tag (25 nM). (3) Detection reagents are added (5 μL of each): anti-His d2 (acceptor) and anti-human IgG terbium (donor) to enable time-resolved FRET between the two proteins when in proximity. (4) After incubation (1 h), the TR-FRET signal is measured as the 665 nm to 620 nm emission ratio, where inhibitor-mediated complex disruption reduces the ratiometric signal relative to vehicle controls.
Materials and reagents
Biological materials
| Reagent | Supplier | Catalog number | Notes |
| Recombinant human SLIT2-His | Sino Biological | 11967-H08H | Aliquot at 10–20 μM. Avoid repeated freeze-thaw cycles; store at -80 °C. Thaw on ice before labeling. Determine protein concentration prior to aliquoting by UV absorbance at 280 nm using the supplier-provided extinction coefficient or Certificate of Analysis values. |
| Recombinant human ROBO1-Fc | Sino Biological | 30073-H02H | Prepare 10–20 μM aliquots; store at -80 °C. Handle gently to prevent aggregation. Determine protein concentration prior to aliquoting by UV absorbance at 280 nm using the supplier-provided extinction coefficient or Certificate of Analysis values. |
Critical note: Glycosylated extracellular proteins like SLIT2 and ROBO1 are prone to aggregation and loss of activity when repeatedly thawed. Prepare small aliquots (20–30 μL) to minimize freeze-thaw events. Before each use, centrifuge briefly (10,000× g, 1 min) to remove any precipitate.
Reagents for TRIC/MST
| Reagent | Supplier | Catalog number | Storage | Purpose/notes |
| RED-tris-NTA 2nd Gen dye | NanoTemper | MO-L018 | -20 °C (light-protected) | His-tag selective labeling reagent for TRIC. Provides a strong signal and minimal aggregation relative to classic NT-647 dyes. |
| HEPES (1 M) | Millipore Sigma | 83264 | RT | Buffering agent; pH 7.4 ensures protein stability and minimizes dye quenching. |
| Tween-20, Ultrapure | Thermo Scientific | J20605.AP | RT | Reduces nonspecific adsorption on plasticware. Use ≤0.005% to avoid interference with TRIC. |
| DMSO | Thermo Scientific | 036480.K2 | RT | Solvent for compounds. Use analytical grade. Maintain identical DMSO across all wells to avoid apparent signal shifts. |
| TargetMol Lipid Metabolism Library | TargetMol | L2510 | -20 °C | 653 compounds; 10 mM in 100% DMSO. Keep plates sealed and equilibrate to RT before opening to avoid condensation. |
| PBS (1×, pH 7.4) | Vendor of choice | - | RT | Buffer option for screening and general protein handling. |
| Sodium chloride (NaCl) | Vendor of choice | - | RT | Used to adjust ionic strength for HEPES-based screening buffer (150 mM NaCl). |
Reagents for TR-FRET
| Reagent | Supplier | Catalog number | Storage | Notes |
| HTRF PAb anti-human IgG-Tb | Revvity/Cisbio | 61HFCTAF or 61HFCTAA | 4 °C (do not freeze) | Donor antibody recognizing ROBO1-Fc. Tb3+ donor ensures long-lifetime fluorescence for time-resolved detection. |
| HTRF Anti-6His mAb d2-Conjugate | Revvity/Cisbio | 61HISDLF | 4 °C | Acceptor fluorophore specific for His-tagged SLIT2. |
| HTRF PPI Tb detection buffer | Revvity/Cisbio | 61DB10RDF | 4 °C | Optimized for HTRF energy transfer. Do not dilute unless otherwise indicated. |
| Bexarotene | Sigma-Aldrich | SML0282-10MG | -20 °C (amber vial) | Positive-control ligand. Prepare 10 mM stock in DMSO; store protected from light. |
Solutions
1. Screening buffer (TRIC/MST) (see Recipes)
2. TR-FRET assay buffer (see Recipes)
Recipes
1. Screening buffer (TRIC/MST)
Screening buffer can be prepared using either (i) 1× PBS (pH 7.4) or (ii) 50 mM HEPES pH 7.4 supplemented with 150 mM NaCl. In both cases, add 0.005% Tween-20. Filter (0.22 μm) and prepare fresh daily. Keep on ice during use.
Note: PBS and HEPES-based buffers were both compatible with this workflow. Maintaining a constant 2% DMSO across all wells reduces viscosity and matrix-driven variability in TRIC. Use minimal detergent (≤0.005% Tween-20) to limit nonspecific adsorption while preserving TRIC sensitivity. If changes are needed for solubility or sticking, adjust one parameter at a time and re-verify baseline stability, signal direction, and replicate variability before screening.
2. TR-FRET assay buffer
Use the supplied PPI Tb detection buffer without further modification. Equilibrate to room temperature before use to prevent condensation on plates.
Laboratory supplies
| Item | Supplier | Catalog number | Notes |
| 384-well barcoded plates (Dianthus) | NanoTemper | DI-P001B | Compatible with automated plate recognition. Use only once to prevent residual dye contamination. |
| White 384-well plates | Greiner | 784075 | Opaque walls maximize FRET signal; avoid transparent plates. |
| Plate seals | Greiner Bio-One | 82050-992 | Ensure tight sealing to prevent evaporation during 1 h incubation. |
| Electronic multichannel pipettor | Integra Viaflo 96 | - | Maintains uniform aspiration speed for accurate small-volume dispensing. |
| Microcentrifuge, vortex mixer | - | - | Routine lab equipment. |
| Low protein-binding microcentrifuge tubes (1.5 mL) | Vendor of choice | - | Recommended for labeled protein handling to minimize adsorption. |
| Monolith capillaries (compatible with Monolith X) | NanoTemper | - | Use the capillaries recommended for Monolith X measurements. |
Equipment
| Instrument | Function/settings |
| NanoTemper Dianthus | For TRIC primary screen, use DI.Control software for QC flags. Typical parameters: IR-laser 100%, MST power medium, 20 min incubation. |
| NanoTemper Monolith X | For affinity determination (Kd). |
| Tecan Infinite M1000 Pro | For TR-FRET detection; excitation = 340 nm, emission = 620 nm and 665 nm, integration delay = 60 μs. |
| Benchtop centrifuge | Used to remove microbubbles and aggregates before TRIC measurement and after labeling clarification. A microcentrifuge rotor compatible with 1.5 mL tubes is sufficient for the labeling clarification spin (15,000× g, 10 min, 4 °C). Quick spins (5–10 s) can be used to clear bubbles before loading capillaries or reading plates. |
Software and datasets
| Software | Use |
| DI.Control (NanoTemper) | TRIC data acquisition and QC flagging (autofluorescence, quenching, aggregation). |
| GraphPad Prism 10 | Nonlinear regression for Kd and IC50, statistical analysis, and graph generation. |
Procedure
文章信息
稿件历史记录
提交日期: Sep 17, 2025
接收日期: Jan 14, 2026
在线发布日期: Jan 30, 2026
出版日期: Feb 20, 2026
版权信息
© 2026 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Gabr, M. T., García-Vázquez, N. and Abdel-Rahman, S. A. (2026). Orthogonal Temperature-Related Intensity Change and Time-Resolved Förster Resonance Energy Transfer High-Throughput Screening Platform for the Discovery of SLIT2 Binders. Bio-protocol 16(4): e5604. DOI: 10.21769/BioProtoc.5604.
分类
药物发现 > 药物筛选
生物化学 > 蛋白质 > 相互作用 > 蛋白质-蛋白质相互作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link