- EN - English
- CN - 中文
Electrophoretic mobility shift assay (EMSA) for assessing RNA-protein binding and complex formation using recombinant RNA-binding proteins and in vitro-transcribed RNA
发布: 2026年05月05日第16卷第9期 DOI: 10.21769/BioProtoc.5583 浏览次数: 168
评审: Anonymous reviewer(s)
Abstract
Evaluating RNA–protein interactions is key to understanding post-transcriptional gene regulation. Electrophoretic mobility shift assays (EMSAs) remain a widely used technique to study these interactions, revealing information about binding affinities and binding modalities, including cooperativity and complex formation. Here, we detail, in a step-by-step protocol, how to perform EMSAs. We describe how to generate, purify, and quantitate 32P-radiolabeled RNA by in vitro transcription, as well as the expression and purification of recombinant RNA-binding proteins in E. coli using ELAV as an example. We then describe how to set up binding reactions using serial dilutions in a microtiter plate format of recombinant ELAV and in vitro–transcribed RNA and how to perform EMSAs using native low-crosslinked acrylamide gels, with detailed graphically supported instructions and troubleshooting guides.
Key features
• Efficient production and purification of radiolabeled RNA probes via in vitro transcription and denaturing PAGE.
• Reproducible binding assays shown using recombinant ELAV protein as an example.
• Quantitative EMSA setup using serial dilutions in a microtiter plate for accurate binding curves.
• Native gel preparation and optimized running conditions for high-resolution separation of RNA–protein complexes.
Keywords: EMSAGraphical overview
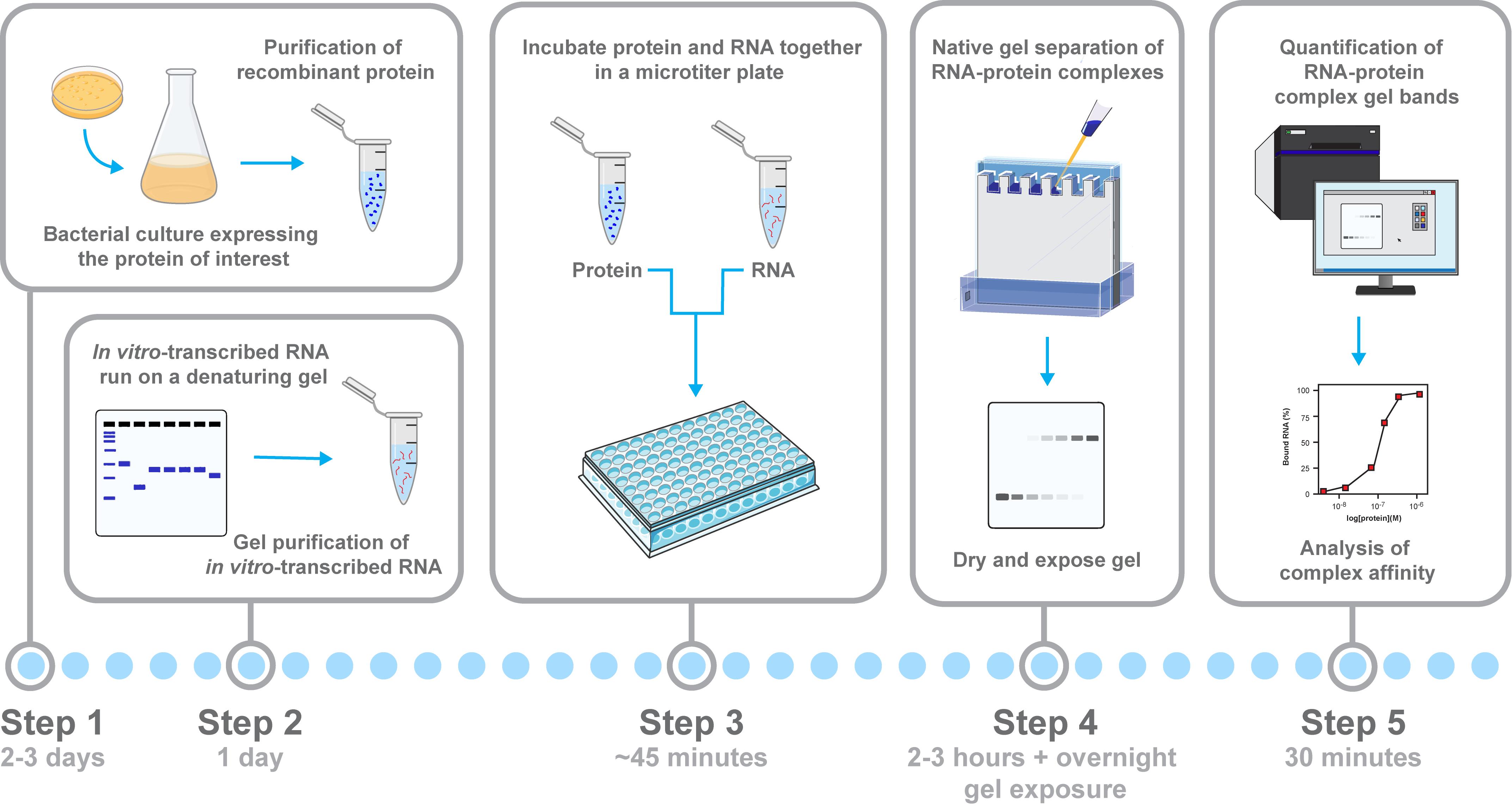
Workflow for assessing RNA–protein binding and complex formation by electrophoretic mobility shift assays (EMSAs). Schematic overview of the workflow for analyzing RNA–protein interactions using an EMSA, with relevant steps highlighted. The recombinant protein is expressed and purified from E. coli, and the 32P-radiolabeled RNA is generated by in vitro transcription and gel-purified. Then, protein serial dilutions are incubated with the RNA probe in a microtiter plate to allow for complex formation. Samples are separated on a native low-crosslinked acrylamide gel, where RNA–protein complexes show reduced mobility compared with free RNA. Key steps highlighted are protein purification, RNA preparation, binding setup, electrophoresis, and signal quantification.
Background
RNA-binding proteins (RBPs) regulate multiple aspects of RNA metabolism, including splicing, polyadenylation, stability, localization, and translation [1–5]. Understanding how RBPs recognize specific RNA sequences or structures is key to uncovering post-transcriptional regulatory mechanisms. Electrophoretic mobility shift assays (EMSAs) remain a gold standard for analyzing direct RNA–protein interactions, as they allow the visualization of complex formation, binding modalities including cooperativity, and determination of binding affinities under near-physiological conditions. In addition, EMSAs are invaluable for mapping nucleic acid binding sites through mutagenesis or identifying key amino acid residues in RBPs that are essential for RNA binding.
The neuronal RBP ELAV (embryonic lethal/abnormal vision) from Drosophila melanogaster, homologous to human Hu family proteins, binds U-rich motifs and plays a critical role in many aspects of mRNA processing, including the inhibition of 3′ end processing leading to extension of 3′ UTRs, alternative splicing, and stabilization, localization, and translation [6–9]. Historically, 32P-radiolabeled RNA probes have been used in EMSAs due to their high sensitivity and easy quantification. Generally, RNA-ELAV/Hu or YTHDC RBP interactions in EMSAs have been highly reproducible [10–15], but the quality of both the RNA and protein components and consistent electrophoresis conditions are important.
This protocol provides instructions to generate and quantify high-purity 32P-radiolabeled RNA via in vitro transcription and gel purification, followed by native EMSAs using the recombinant ELAV protein. It includes detailed steps for gel preparation, binding reaction setup in a microtiter format, and critical handling of glass plates and buffers. The protocol is adaptable to other RBPs and can be used for other assays analyzing RNA–protein interactions, including footprinting assays, minimal binding site mapping, or analysis of large complexes [11,15–17]. We further provide a guide for troubleshooting and quantification of binding and complex assembly characteristics.
Materials and reagents
Biological materials
1. Escherichia coli strain BL21(DE3) (e.g., NEB, C2527I) for the expression of GST- ELAV
Reagents
1. Tryptone (e.g., MilliporeSigma, catalog number: 16922)
2. Yeast extract (MilliporeSigma, catalog number: 70161)
3. Ampicillin (e.g., Sigma-Aldrich, catalog number: A9518)
4. Isopropyl β-D-1-thiogalactopyranoside (IPTG) (e.g., Sigma-Aldrich, catalog number: I6758)
5. Phosphate-buffered saline (PBS+), DEPC-treated and supplemented with 1 mM EDTA, 1 mM DTT, 5 μg/mL leupeptin prepared with DEPC-treated water, and 1 mM phenylmethylsulphonyl fluoride (PMSF) from 200 mM stock in isopropanol
6. Lysozyme (e.g., Sigma-Aldrich, catalog number: L7651)
7. Glutathione Sepharose 4B (e.g., Cytiva, catalog number: 17-0756-01)
8. NP-40/Igepal CA-630 (e.g., Sigma-Aldrich, catalog number: I3021)
9. PreScission protease (e.g., Cytiva/Amersham, catalog number: GE27-0843-01)
10. Protease inhibitor cocktail (Roche, Merck, catalog number: 4693116001)
11. High-quality glycerol (Invitrogen, catalog number: 1150874)
12. Diethyl pyrocarbonate (DEPC) to make RNase-free water (e.g., Thermo Fisher, catalog number: 10977015)
13. Liquid nitrogen
14. Phenol:chloroform:isoamyl alcohol (25:24:1) (e.g., Thermo Fisher, catalog number: 15593031)
15. Sodium acetate, pH 5.2 (e.g., Thermo Fisher, catalog number: AM9740)
16. Nucleotide mix (cold and hot NTPs, e.g., to label with α-32P ATP, ATP 0.2 mM, UTP 10 mM, GTP 10 mM, CTP 10 mM, final concentrations adjustable)
17. 10× transcription buffer (commercially supplied with T3/T7/SP6 polymerases)
18. Radiolabeled nucleotides (e.g., α-32P ATP or UTP, 800 Ci/mmol, 10 miCi/mL, 12.5 μM, PerkinElmer or Hartmann Analytics)
CAUTION: When handling radioactive nucleotides, always follow institutional radiation safety regulations.
19. RNasin Plus RNase inhibitor (e.g., Promega, catalog number: N2615)
20. RNA polymerase (T3, T7, or SP6) (Ambion)
21. DNase I (Ambion, catalog number: AM2222)
22. Dithiothreitol (DTT) (e.g., Thermo Fisher, catalog number: R0861)
23. Ethylenediaminetetraacetic acid (EDTA) (e.g., Thermo Fisher, catalog number: 15575020)
24. Ethanol (molecular biology grade) (e.g., Sigma-Aldrich, catalog number: E7023)
25. HEPES, pH 7.5 (e.g., Sigma-Aldrich, catalog number: H4034)
26. NaCl (e.g., Sigma-Aldrich, catalog number: S7653)
27. 2-Mercaptoethanol (optional, if used in sample buffers) (e.g., Sigma-Aldrich, catalog number: M6250)
28. Rain-X (Amazon) to silanize the inner plate for denaturing gels
29. Dishwashing liquid soap and 1 M KOH in methanol to wash EMSA plates
30. Tetramethylethylenediamine (TEMED) (e.g., Bio-Rad, catalog number: 1610801)
31. Ammonium persulfate (APS) (e.g., Sigma-Aldrich, catalog number: A3678)
32. Formamide (deionized) (e.g., Thermo Fisher, catalog number: AM9342)
33. Bromophenol blue (e.g., Sigma-Aldrich, catalog number: B0126)
34. Xylene cyanol FF (e.g., Thermo Fisher, catalog number: BP125)
35. Acetylated bovine serum albumin (BSA) (e.g., Thermo Fisher, catalog number: A2427)
36. Complete Mini EDTA-free protease inhibitor cocktail (e.g., Roche, catalog number: 11836170001)
37. pBluescript or pUC19 plasmid with RNA target sequence cloned downstream of T7/T3/SP6 promoter; pUC19 is used as a positive control in the NEB Gibson cloning kit (NEB, catalog number: E5520S)
38. Sodium dodecyl sulfate (SDS) (e.g., Thermo Fisher, catalog number: BP166)
39. TBE buffer, 10× or 5× (e.g., Thermo Fisher, catalog number: BP1333)
40. Tris-HCl, pH 7.5 (e.g., Sigma-Aldrich, catalog number: T5941)
41. tRNA (yeast, 10 mg/mL) (e.g., Sigma-Aldrich, catalog number: R5636)
42. 100 g acrylamide bottle (Bio-Rad, catalog number: 1610100)
43. Bis-acrylamide solution (Bio-Rad, catalog number: 1610142)
Optional reagents
1. Faustovirus capping enzyme (NEB, catalog number: M2081S)
2. Di-nucleotide cap (JENA Bioscience, catalog number: NU-854S)
Solutions
1. LB (Luria-Bertani) media (see Recipes)
2. LB agar (see Recipes)
3. CV buffer (cleavage buffer) (see Recipes)
4. Acrylamide/Bis-acrylamide solution (80:1) (see Recipes)
5. Formamide loading dye (blue juice) (see Recipes)
6. Cracking buffer (see Recipes)
7. Buffer A (see Recipes)
8. Buffer B (see Recipes)
Recipes
1. LB media (per liter)
10 g of tryptone
5 g of yeast extract
10 g of NaCl
2. LB agar (per liter)
LB media plus 15 g of agar
3. CV buffer (cleavage buffer)
50 mM Tris or HEPES pH 7.5 (at 25 °C)
150 mM NaCl
1 mM EDTA
1 mM DTT
4. Acrylamide/Bis-acrylamide solution (80:1)
In a 100 g acrylamide bottle, add 62.5 mL of 2% bis-acrylamide solution (or 1.25 g of bis-acrylamide powder if used) and fill up to 250 mL with DEPC water. Use a new 50 mL Falcon tube to measure the volume, using its grading line on top (the grading on the side is not correct). Avoid acrylamide powder in the air, as acrylamide is highly toxic.
5. Formamide loading dye (blue juice)
Add deionized formamide to 10 mg/mL bromophenol blue, 10 mg/mL xylene cyanol FF, and 5 mM EDTA.
6. Cracking buffer
Add 0.3 M sodium acetate (pH 5.2) in DEPC-treated water and add 0.2% SDS from the 20% stock.
7. Buffer A
Add 400 mM Tris or HEPES pH 7.5 (at 25 °C), 450 mM NaCl, 3 mM EDTA, 3 mM DTT, 0.25 mg/mL tRNA, and 0.5 mg/mL acetylated BSA.
8. Buffer B
Add 300 μL of CV buffer, 150 μL of 5× buffer A, and 300 μL of H2O (150 mM NaCl final).
Equipment
1. Shaking incubator (e.g., Innova 44, New Brunswick)
2. Centrifuge (e.g., Sigma 3-30KS with swing-out rotor, 11134)
3. Microtip sonicator (XL, Heart Systems, model: XL2020)
4. Syringe and 0.45 μM filter (Millipore)
5. Perpex peristaltic pump, including tubing and connectors
6. Amersham Typhoon biomolecular imager (Cytiva or equivalent)
7. Scintillation counter (Beckmann or equivalent)
8. Gel tank (custom-made) and glass plates (20 cm × 20 cm inner plate, 20 cm × 22 cm outer plate) (e.g., Bio-Rad)
9. Spacers and combs (0.75 mm spacers and 20-well combs for EMSA, 1.5 mm spacers and 10-well combs for denaturing gel purification, 0.3 mm spacers and 20-well combs for analytical denaturing gel) (Bio-Rad)
10. Blu-tack (Amazon) to seal plates and tank
11. Powerpacks 200 V for EMSAs and 3,000 V for denaturing gels
12. EMSA gel loading tips (Alphalab, catalog number: LW1100R, 1–200 μL)
13. Denaturing gel loading tips (Corning, gel-loading pipette tips, catalog number: CLS4884, 1–200 μL, end of tip thickness 0.2 mm, tip flat)
14. Nanodrop or equivalent
15. 96-well clear V-bottom TC-treated microplate (Corning, catalog number: 3894)
Software and datasets
1. Quantity One 1-D Analysis Software (Bio-Rad) or equivalent (e.g., ImageJ)
Procedure
文章信息
稿件历史记录
提交日期: Nov 3, 2025
接收日期: Dec 23, 2025
在线发布日期: Jan 8, 2026
出版日期: May 5, 2026
版权信息
© 2026 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
McQuarrie, D. W. J. and Soller, M. (2026). Electrophoretic mobility shift assay (EMSA) for assessing RNA-protein binding and complex formation using recombinant RNA-binding proteins and in vitro-transcribed RNA. Bio-protocol 16(9): e5583. DOI: 10.21769/BioProtoc.5583.
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link









