- EN - English
- CN - 中文
High Content In Vitro Survival Assay of Cortical Neurons
皮层神经元的高内涵体外存活检测方法
(*contributed equally to this work) 发布: 2026年02月05日第16卷第3期 DOI: 10.21769/BioProtoc.5582 浏览次数: 132
评审: Anonymous reviewer(s)
Abstract
Neuronal survival in vitro is usually used as a parameter to assess the effect of drug treatments or genetic manipulation in a disease condition. Easy and inexpensive protocols based on neuronal metabolism, such as 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), provide a global view of protective or toxic effects but do not allow for the monitoring of cell survival at the single neuronal level over time. By utilizing live imaging microscopy with a high-throughput microscope, we monitored transduced primary cortical neurons from 7–21 days in vitro (DIV) at the single neuronal level. We established a semi-automated analysis pipeline that incorporates data stratification to minimize the misleading impact of neuronal trophic effects due to plating variability; here, we provide all the necessary commands to reproduce it.
Key features
• The protocol enables monitoring of primary cortical neuron survival from DIV 7 to 21 in 96-well plates following various cellular treatments.
• It provides single-cell and real-time imaging resolution, enabling the identification of small changes in viability over time.
• It provides a detailed description of semi-automated neuronal detection over time.
• It relies on data stratification based on the neuronal starting number, which helps reduce the impact of neuronal trophic effects due to plating variability.
• It has been used to assess the effect of glial extracellular vesicles on cortical neurons, as reported in [1].
Keywords: Neuronal survival (神经元存活)Graphical overview
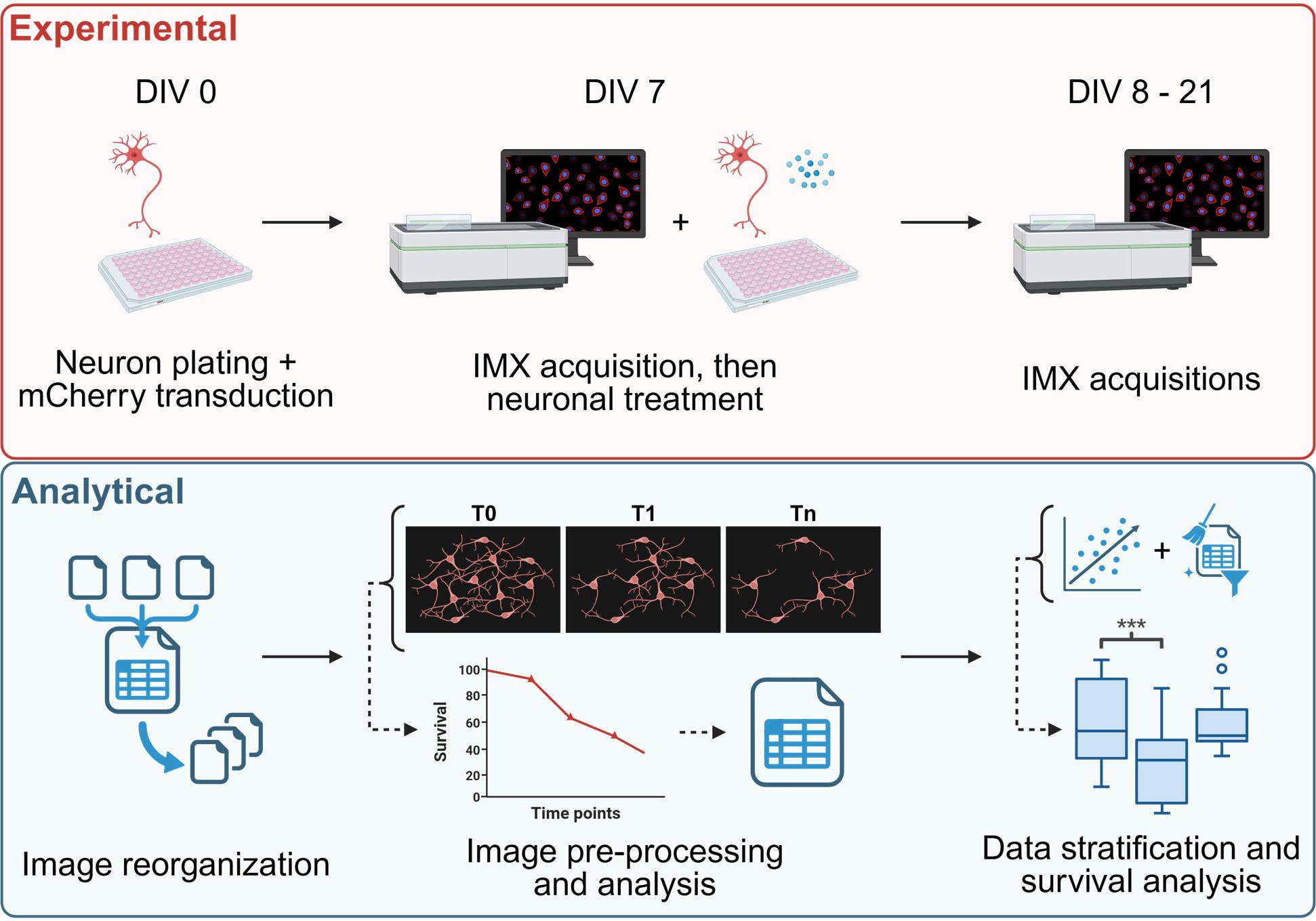
High content survival assay of cortical murine primary neurons, allowing data stratification to account for the trophic effects. IMX= ImageXpress®.
Background
Survival assays measure how treatments affect neuronal viability, but primary cultures are challenging because plating variability affects the number of surviving neurons. However, by taking advantage of their low mobility, we report an innovative, high-content, in vitro survival assay that can image and monitor primary neuronal culture over two weeks. By integrating live imaging and stratifying data by the initial neuronal count, the assay overcomes plating-related variability, including trophic effects, as detailed in our recent publication [1].
Most of the current methods used to quantify the survival of primary cultures require the neuronal cultures to be interrupted at the selected time point. For example, to perform an ELISA assay [2] or any immunofluorescence assay, the cells must be fixed. MTT and luciferase assays (the latter for neurons transfected with firefly luciferase reporter) require cell lysis to quantify survival rates [3]. Trypan blue and propidium iodide staining are membrane permeability-based assays that require cell detachment for counting [3].
Analyses that use culture media as the starting material, such as the quantification of lactate dehydrogenase (LDH) released [3], do not require cell cultures to be lysed or fixed; however, they are unable to account for disparities induced by plating variability and the relative neuronal trophic effect.
A neuronal survival assay in primary neuronal culture, utilizing single-cell and real-time imaging resolution, has been previously reported [4]. Similarly, our method relies on live imaging of fluorescently transduced neurons, enabling the comparison of different time points within the same neuronal culture. Additionally, it is combined with post-acquisition data stratification based on the initial cell number.
Although our novel method has clear advantages over previously published survival assays, some technical limitations are present. First, neuronal primary cultures are sensitive to temperature and CO2 changes induced by plate relocation during the incubator-microscope shift, so this must be considered. For this reason, daily acquisitions over a long period (more than five days) are not recommended. Additionally, technical difficulties may arise when attempting to minimize the shift of the field of view (FOV) during acquisitions at different time points. The position of the plate inside the ImageXpress is crucial, and re-aligning the FOV with the previous ones may take time. Furthermore, the analysis parameters (both in image preprocessing, processing, and post-analysis stratification) must be set up depending on the conditions used for the experiment (e.g., the type of cell used, the selected cell treatment, etc.). In conclusion, it is essential to emphasize that the overexpression of fluorescent reporter proteins through lentiviral transduction results in a slight increase in cell mortality compared to non-transduced controls.
Our survival assay was specifically designed for primary murine cortical neurons, but it can be applied to other primary neuronal cultures or to any other cell culture in which cells have low migration and have exited the cell cycle, such as post-mitotic cells. Through this protocol, neuronal survival curves can be calculated at specific selected time points, allowing several analytical approaches; the conditions of interest, such as enhancement or suppression of biological pathways through drug treatment or genetic modifications, can be followed intra-condition during all time points, or be compared inter-condition with the relative controls at each specific time point.
As previously reported, the aim of this protocol is to conduct a quantitative analysis of neuronal survival with or without specific treatments. The first step involves producing lentiviral particles to induce expression of the fluorescent reporter protein in neurons. Once the viral particles have been isolated from HEK293T cells and quantified, the murine cortical neurons can be plated and transduced. After seven days in culture, the first live imaging acquisition is performed, and the neurons are treated as desired. Additional acquisitions will be performed at different time points (three to five additional time points are recommended). All the images are then reorganized, preprocessed, and analyzed using a semi-automatic Fiji macro pipeline, resulting in a CSV table containing the neuronal count and relative survival rate for each field of view at each time point. Starting from the treatment control, the neuronal trophic effect can then be established, and specific data stratification can be applied to rule out its influence on the survival rate. Once the stratification is optimized, quantitative comparisons of interest can be performed.
To design the experiment properly, it is essential to define the number of conditions to be compared, identify the necessary controls, and determine the number of technical and biological replicates required. Once all of these are acknowledged, the number of wells (and so the number of plates) that are necessary for the experiment can be calculated, as well as the relative quantity of lentiviral particles needed for the transduction. During neuronal plating, it is essential to avoid cellular overconfluency; therefore, we suggest plating 15,000 cells/well in a 96-well plate.
Materials and reagents
Biological materials
Plasmids
1. psPAX2 (Addgene #12260): plasmid containing gag, pol, and rev genes
2. pMD2.G (Addgene #12259): plasmid containing VSV-G envelope gene
3. LentiLox 3.7 syn-mCherry: mCherry sequence under the control of the neuron-specific synapsin I promoter was cloned into a LentiLox 3.7 vector [5]; this newly generated plasmid is available upon request
Cell lines
1. Murine primary cortical neuronal cultures [obtained from embryos (E15.5) derived from wild-type C57BL/6N mice (Charles River Laboratories, ITALIA)]
2. HEK293T cells [American Type Culture Collection (ATCC), catalog number: CRL-3216, origin: human epithelial-like cells isolated from the kidney]
Primers
1. Forward primer: TCCTGCTCAACTTCCTGTCGAG
2. Reverse primer: CACAGGTCAAACCTCCTAGGAATG
Note: 100 μM desalted primers can be purchased from any company that provides this service (e.g., Eurofins, Macrogen).
Reagents
Murine primary cortical neuronal cultures
1. Minimum essential medium (MEM), GlutaMAXTM supplement (Life Tech., catalog number: 41090-028)
2. Fetal bovine serum (FBS) (Gibco, catalog number: 10270-106)
3. Neurobasal (Life Tech., catalog number: 21103-049)
4. Sodium pyruvate (Invitrogen, catalog number: 11360-039)
5. Cytosine arabinoside (AraC) (100 mM) (Merck, catalog number: C1768)
6. Penicillin-streptomycin (PenStrep) (Gibco, catalog number: 15140-122)
7. B27 (50×) (Gibco, catalog number: 17504-044)
8. Poly-D-lysine (Merck, catalog number: P7280)
9. Papain (Merck, catalog number: P4762)
10. Sodium acetate (Merck, catalog number: 241245)
11. DNase I (Merck, catalog number: D5025)
12. L-Cystine (Merck, catalog number: C7602)
13. 1 M hydrochloric acid (HCl) (Merck, catalog number: H9892)
14. Earle’s balanced salt solution (EBSS) (10×) (Merck, catalog number: E7510)
15. Sodium bicarbonate (Merck, catalog number: S5761)
16. Trypsin inhibitor (Merck, catalog number: T9253)
17. Bovine serum albumin (Merck, catalog number: A7030)
18. Ethylenediaminetetra acetic acid (EDTA) (Invitrogen, catalog number: 15575-038)
19. L-Glutamine (Gibco, catalog number: 25030-081)
20. 0.22 μm filters (Euroclone, catalog number: EPSPV2230)
HEK293T cultures and lentiviral particles production and quantification
1. Dulbecco’s modified Eagle medium (DMEM) 1× (Gibco, catalog number: 11960-044)
2. Fetal bovine serum (FBS) (Gibco, catalog number: 10270-106)
3. Penicillin-streptomycin (PenStrep) (Gibco, catalog number: 15140-122)
4. L-Glutamine (Gibco, catalog number: 25030-081)
5. Sodium chloride (NaCl) (Merck, catalog number: S9888)
6. HEPES (N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid), 0.5 M buffer solution, pH 7.0 (Thermo Fisher Scientific, catalog number: J60064.AE)
7. Disodium phosphate (Na2HPO4) (Merck, catalog number: S9763)
8. Calcium chloride (CaCl2) (Merck, catalog number: C5670)
9. 0.22 μm filters (Euroclone, catalog number: EPSPV2230)
10. Triton X-100 (Merck, catalog number: T8532)
11. Potassium chloride (KCl) (Merck, catalog number: P3911)
12. Tris base (Merck, catalog number: T6687)
13. Glycerol (Merck, catalog number: G5516)
14. Ribolock RNase inhibitor (Thermo Fisher Scientific, catalog number: EO0381)
15. Ammonium sulfate [(NH4)2SO4] (Merck, catalog number: A4915)
16. Magnesium chloride (MgCl2) (Thermo Fisher Scientific, catalog number: R0971)
17. BSA (NEB, catalog number: B9000)
18. SYBR Green I (Thermo Fisher Scientific, catalog number: S7563)
19. Tris-EDTA (TE) buffer solution, pH 8.0, RNase-free (Invitrogen, catalog number: AM9849)
20. dNTPs (Thermo Fisher Scientific, catalog number: R0191)
21. MS2 RNA (Roche, catalog number: 10165948001)
22. DreamTaq Hot Start DNA Polymerase (Thermo Fisher Scientific, catalog number: EP1701)
Additional materials
1. Ethanol (Merck, catalog number: 459844)
2. PhenoPlate 96-well (PerkinElmer, catalog number: 6055300)
Solutions
1. Sodium acetate solution pH 4.5 (50 mM) (see Recipes)
2. Papain stock solution (see Recipes)
3. DNase I stock solution (see Recipes)
4. L-Cystine stock solution (see Recipes)
5. 1× EBSS solution (see Recipes)
6. 10/10 solution (see Recipes)
7. Papain solution (see Recipes)
8. EBSS-10/10 solution (see Recipes)
9. Plating media (see Recipes)
10. Neurobasal complete (see Recipes)
11. 1× Poly-D-lysine solution (0.01 mg/mL) (see Recipes)
12. DMEM complete (see Recipes)
13. NaCl stock solution (5 M) (see Recipes)
14. Na2HPO4 stock solution (0.15 M) (see Recipes)
15. 2× HPB (see Recipes)
16. CaCl2 stock solution (2.5 M) (see Recipes)
17. KCl stock solution (5 M) (see Recipes)
18. Tris-HCl pH 7.4 (1 M) and pH 8.3 (2 M) (see Recipes)
19. 2× lysis buffer (see Recipes)
20. (NH4)2SO4 stock solution (500 mM) (see Recipes)
21. 10× core buffer (see Recipes)
22. MgCl2 stock solution (25 mM) (see Recipes)
23. SYBR Green I stock solution (see Recipes)
24. MS2 RNA stock solution (see Recipes)
25. 2× reaction buffer (see Recipes)
Recipes
1. Sodium acetate solution pH 4.5 (50 mM)
Prepare an aqueous solution of sodium acetate and adjust it to the appropriate pH with a solution of HCl. Prepare the solution fresh and keep it at room temperature (RT) before use.
2. Papain stock solution
Dissolve lyophilized papain powder in 50 mM sodium acetate, pH 4.5 (prepared from the stock solution), to a stock concentration of 100 units/mL. Store at 4 °C for up to 6 months.
3. DNase I stock solution
Dissolve lyophilized DNase I powder in sterile water to prepare a stock solution of 1 mg/mL. Store at -20 °C for up to 6 months.
4. L-Cystine stock solution
Dissolve L-Cystine in 1 M HCl to prepare a stock solution of 100 mM. Store at 4 °C for up to 6 months.
5. 1× EBSS solution (for 500 mL)
Add 50 mL of 10× EBSS and 1.1 g of sodium bicarbonate. Adjust the volume with double-distilled water and filter-sterilize. Store at 4 °C for up to 1 month.
6. 10/10 solution (for 50 mL)
Add 500 mg of trypsin inhibitor and 500 mg of bovine serum albumin (Merck, A7030) and adjust the volume with 1× EBSS solution. Filter-sterilize and store at 4 °C for up to 1 month.
7. Papain solution (for 30 mL)
Add 200 μL of papain stock solution, 30 μL of EDTA, and 30 μL of cysteine stock solution (100 mM). Adjust the volume with 1× EBSS solution and filter-sterilize. Prepare fresh and keep at 37 °C before use.
8. EBSS-10/10 solution
Add 4.5 mL of 1× EBSS solution and 0.5 mL of 10/10 solution. Prepare fresh and keep at 37 °C before use.
9. Plating media (for 50 mL)
Add 5 mL of FBS and 0.5 mL of PenStrep and adjust the volume with MEM, GlutaMAXTM supplement. Prepare fresh and keep at 37 °C before use.
10. Neurobasal complete (for 50 mL)
Add 0.5 mL of sodium pyruvate, 1 mL of B27 (50×), 0.5 mL of PenStrep, 0.5 mL of L-Glutamine, and 50 μL of Ara C (100 mM). Adjust the volume with neurobasal medium. Prepare fresh and keep at 37 °C before use.
11. 1× Poly-D-lysine solution (0.01 mg/mL)
Dilute the 1 mg/mL Poly-D-lysine stock solution 1:100 in PBS 1×. Prepare fresh and keep at RT before use.
12. DMEM complete
Supplement DMEM 1× with 10% FBS, 1% PenStrep, and 1% L-Glutamine. Store at 4 °C for up to 1 month.
13. NaCl stock solution (5 M)
Prepare an aqueous solution of NaCl at the appropriate concentration. Store at RT for up to 6 months.
14. Na2HPO4 stock solution (0.15 M)
Prepare an aqueous solution of Na2HPO4 at the appropriate concentration. Store at RT for up to 6 months.
15. 2× HPB (for 100 mL)
Add 0.28 M NaCl (5.6 mL of 5 M NaCl stock solution), 0.05 M HEPES pH 7.0, and 0.0015 M Na2HPO4 (1 mL of 0.15 M Na2HPO4 stock solution). Adjust pH to 7.1 with NaOH. Adjust the volume with double-distilled water. Filter-sterilize and store at 4 °C for up to 6 months.
16. CaCl2 stock solution (2.5 M)
Prepare an aqueous solution of CaCl2 at the appropriate concentration. Filter-sterilize and store at 4 °C for up to 6 months.
17. KCl stock solution (5 M)
Prepare an aqueous solution of KCl at the appropriate concentration. Store at RT for up to 6 months.
18. Tris-HCl pH 7.4 (1 M) and pH 8.3 (2 M)
Prepare an aqueous solution of Tris base and adjust it to the appropriate pH with a solution of HCl. Store at RT for up to 6 months.
19. 2× lysis buffer
Add 0.25% Triton X-100, 50 mM KCl (from stock solution), 100 mM Tris-HCl, pH 7.4 (from stock solution), 40% glycerol, and 0.8 U/μL Ribolock RNase inhibitor. This buffer can be stored at RT for up to 6 months without RNase inhibitor. Prepare a freshly aliquoted complete buffer from the incomplete one and add the appropriate amount of RNase inhibitor just before viral particle lysis.
20. (NH4)2SO4 stock solution (500 mM)
Prepare an aqueous solution of (NH4)2SO4 at the appropriate concentration. Store at RT for up to 6 months.
21. 10× core buffer
Add 50 mM (NH4)2SO4 (from stock solution), 200 mM KCl (from stock solution), and 200 mM Tris-HCl, pH 8.3 (from stock solution). Adjust the volume with double-distilled water. Store at RT for up to 6 months.
22. MgCl2 stock solution (25 mM)
Prepare an aqueous solution of MgCl2 at the appropriate concentration. Store at RT for up to 6 months.
23. SYBR Green I stock solution
Dilute the original SYBR Green I stock 1:100 in TE buffer solution, pH 8.0, RNase-free. Aliquot (20 μL) and store at -80 °C for up to 6 months. Once thawed, do not reuse.
24. MS2 RNA stock solution
The original MS2 RNA stock is 0.8 μg/µL, i.e., 0.7 pmol/µL. Aliquot (15 μL) and store at -80 °C for up to 6 months.
25. 2× reaction buffer
Add 5 mM (NH4)2SO4, 20 mM KCl, 20 mM Tris-HCl (pH 8.3), 10 mM MgCl2, 0.2 mg/mL BSA, 1:10,000 SYBR Green I, 400 μM dNTPs, 1 μM forward primer, 1 μM reverse primer, 7 pmol/mL MS2 RNA, and 20 U of RNase inhibitors. Prepare using a 10× core buffer according to the table below and store in 50 μL aliquots at -80 °C for up to 6 months.
| Compound | Stock | 2 reactions (μL) | 100 reactions (μL) | 300 reactions (μL) |
|---|---|---|---|---|
| 10× core buffer | Stock solution | 1 | 50 | 150 |
| MgCl2 25 mM | Stock solution | 4 | 200 | 600 |
| 100× BSA | NEB, B9000 | 0.2 | 10 | 30 |
| dNTPs 10 mM | Thermo Fisher Scientific, R0191 | 0.4 | 20 | 60 |
| Primer fwd 100 μM | Stock solution | 0.1 | 5 | 15 |
| Primer rev 100 μM | Stock solution | 0.1 | 5 | 15 |
| MS2 RNA | Roche, 10165948001 | 0.1 | 5 | 15 |
| SYBR Green I 1:100 | Stock solution | 0.1 | 5 | 15 |
| H2O | 4 | 200 | 600 | |
| RNase inhibitor | Thermo Fisher Scientific, EO0381 | 0.01 | 0.5 | 1.5 |
Equipment
1. ImageXpress® micro confocal high-content inverted imaging system from Molecular Devices, equipped with:
a. Andor Zyla 4.2 Megapixel sCMOS digital camera
b. Lumencor Spectra light engine LED light source
c. 576/23 nm excitation filter, 624/40 nm emission filter, and 594 nm single-band dichroic mirror
d. Nikon Plan Apo 10×/0.45 dry objective
e. Incubation system with temperature and CO2 control
2. Tabletop centrifuge (Eppendorf, model: Centrifuge 5804)
3. Touch Real-Time PCR Detection System (Bio-Rad, model: CFX96)
Software and datasets
1. MetaXpress from Molecular Devices, for image acquisition (https://it.moleculardevices.com/products/cellular-imaging-systems/high-content-analysis/metaxpress)
2. Python 3.13, for microscopy data organization (https://www.python.org/downloads/)
3. Fiji (Fiji Is Just ImageJ), for image analysis (https://imagej.net/software/fiji/downloads)
4. GraphPad Prism, for data representation (https://www.graphpad.com/features)
Programming languages
1. ImageJ macro language
2. Jython
3. Python3
Procedure
文章信息
稿件历史记录
提交日期: Oct 28, 2025
接收日期: Dec 18, 2025
在线发布日期: Jan 8, 2026
出版日期: Feb 5, 2026
版权信息
© 2026 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Fioretti, P. V., Roccuzzo, M., Saccon, E., Pennuto, M. and Basso, M. (2026). High Content In Vitro Survival Assay of Cortical Neurons. Bio-protocol 16(3): e5582. DOI: 10.21769/BioProtoc.5582.
分类
神经科学 > 基础技术 > 高通量筛选
细胞生物学 > 细胞活力 > 细胞存活
细胞生物学 > 细胞成像 > 活细胞成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











