- EN - English
- CN - 中文
Quantifying Mechanical Strain–Induced Membrane Damage in Early Neuronal Cells Using an In Vitro Traumatic Brain Injury Model
基于体外创伤性脑损伤模型定量分析机械应变诱导的早期神经元细胞膜损伤
发布: 2026年02月05日第16卷第3期 DOI: 10.21769/BioProtoc.5580 浏览次数: 115
评审: Sébastien GillotinAnonymous reviewer(s)

相关实验方案

基于 rAAV-α-Syn 与 α-Syn 预成纤维共同构建的帕金森病一体化小鼠模型
Santhosh Kumar Subramanya [...] Poonam Thakur
2025年12月05日 1660 阅读
Abstract
This protocol describes a reproducible workflow for modeling in vitro impact-induced traumatic brain injury (TBI) using a mechanical stretch system applied to differentiated SH-SY5Y human neuroblastoma cells cultured on polydimethylsiloxane (PDMS) substrates. The protocol integrates three primary components: (1) fabrication and surface modification of deformable PDMS chambers to support cellular adhesion, (2) partial differentiation of SH-SY5Y cells using retinoic acid, and (3) induction of controlled mechanical strain to simulate mild to moderate TBI. The stretch-induced injury model enables quantitative assessment of cellular viability and recovery following mechanical insult. This approach provides a versatile platform for studying cellular and molecular mechanisms of TBI, screening neuroprotective compounds, and exploring mechanobiological responses in neural cells under controlled strain magnitudes and rates.
Key features
• Provides reproducible in vitro modeling of traumatic brain injury in differentiated SH-SY5Y neurons via controlled mechanical stretch.
• Combines PDMS chamber fabrication and polydopamine coating to generate deformable, biocompatible, neuron-adhesive substrates for mechanobiology assays.
• Flexible platform for testing neuroprotective compounds and investigating mechanotransduction pathways under physiologically relevant stretch conditions.
Keywords: Mechanobiology (力学生物学)Graphical overview
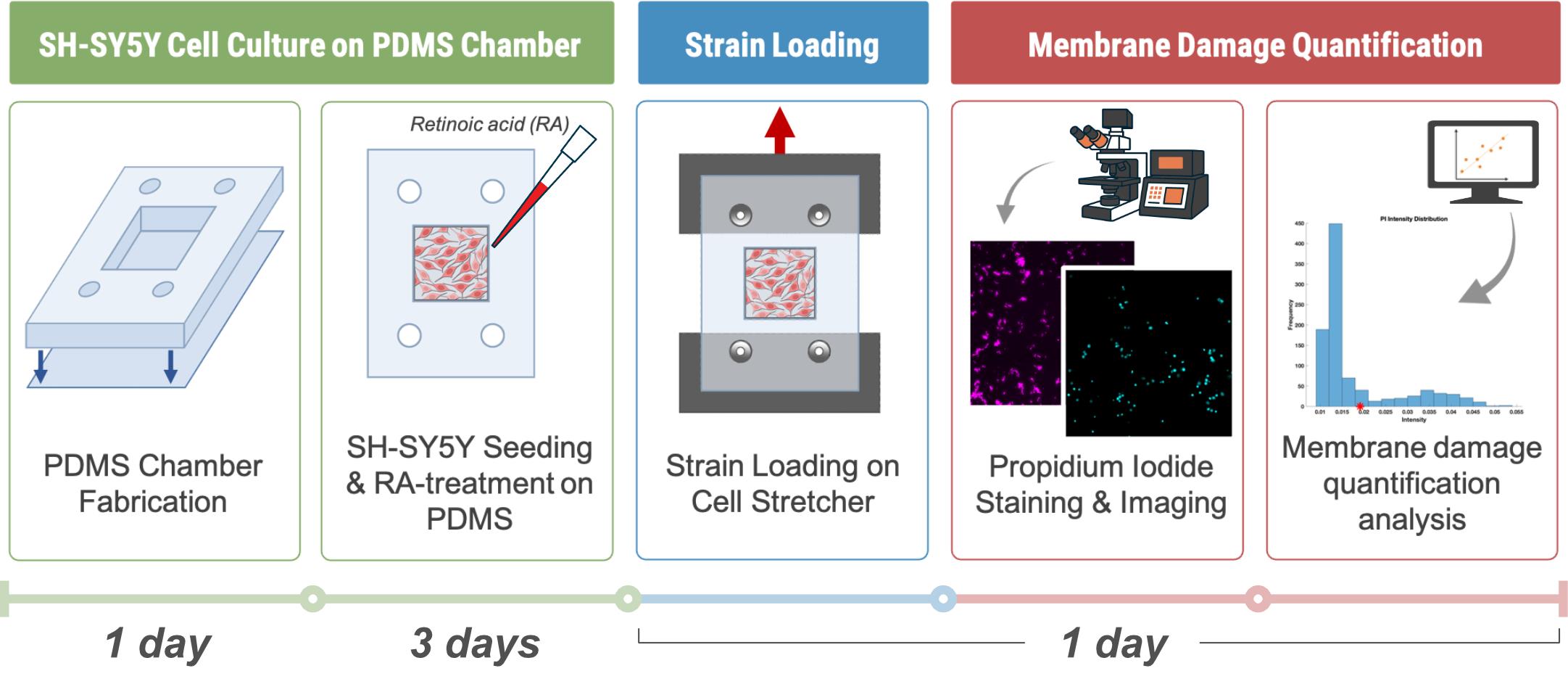
Background
Traumatic brain injury (TBI) is a leading cause of long-term neurological disability and mortality worldwide [1]. Conventional in vivo models of TBI provide valuable physiological insights but are often limited by biological complexity, ethical constraints, and difficulty in isolating specific injury parameters. In contrast, in vitro cell stretching systems offer a controlled, reproducible, and cost-effective platform to study neuronal response to mechanical injury at the cellular level. The human SH-SY5Y neuroblastoma cell line offers a human-derived in vitro model for studying neuronal cytoskeletal responses [2–5]. While they are not fully representative of mature neurons, SH-SY5Y cells may be treated to induce differentiation into neuron-like cells that exhibit neurite outgrowth and express neuronal markers. These cells can be cultured on deformable substrates, such as wells made from polydimethylsiloxane (PDMS), which allow for controlled mechanical deformation. PDMS is biocompatible, transparent, and elastomeric, making it well-suited for modeling brain tissue deformation [6]. However, native PDMS surfaces are hydrophobic [7], necessitating surface functionalization to support neuronal growth. Polydopamine (PDA) coatings, inspired by mussel adhesion chemistry [8,9], provide a simple and effective means of improving PDMS surface hydrophilicity and cell adhesion. In this protocol, PDMS chambers are fabricated and coated with PDA to create a biocompatible substrate for SH-SY5Y culture and differentiation. Differentiation is induced using retinoic acid (RA), yielding cells with extended neurites. Once differentiation is complete, the PDMS substrates are subjected to controlled mechanical stretching to simulate TBI conditions. By adjusting strain magnitude and strain rate, this system allows precise modulation of injury severity and can be combined with a high-resolution optical microscope to examine cellular responses to mechanical injury.
Materials and reagents
Biological materials
1. SH-SY5Y human neuroblastoma cells (ATCC CRL-2266)
Reagents
1. Tris-HCl (MilliporeSigma, catalog number: 648317100GM)
2. Dopamine hydrochloride, powder ≥98% (Sigma-Aldrich, catalog number: H8502-5G)
3. Retinoic acid (RA), all-trans (Sigma-Aldrich, catalog number: R2625)
4. 95% ethanol (Commercial Alcohols, catalog number: P016EA95)
5. Fetal bovine serum (FBS), heat-inactivated (Thermo Fisher Scientific, catalog number: 10270106)
6. Dulbecco’s modified Eagle’s medium (DMEM) with 4.5 g/L D-Glucose and phenol red (Gibco, catalog number: 11965118)
7. Penicillin-streptomycin 100× solution (Cytiva, catalog number: SV30010)
8. Phosphate-buffered saline (PBS), 1× with calcium and magnesium (Corning, catalog number: 21-030-CV)
9. Trypsin-EDTA solution, 0.25% (Gibco, catalog number: 25200056)
10. SYLGARDTM 184 Silicone Elastomer kit (Dow Corning, catalog number: 184 SIL ELAST KIT)
11. Gel-Pak PDMS sheets (PF-60x60-0065-X4)
12. Hydrochloric acid (HCl), 12 N (Ricca Chemical Company, catalog number: R37800001A)
13. Hoechst (Abcam, catalog number: ab228551)
14. Propidium iodide (PI) (BioShop Canada, catalog number: PPI888.5)
Solutions
1. Tris-HCl buffer (see Recipes)
2. Polydopamine (PDA) solution (see Recipes)
3. Retinoic acid (RA) stock solution (see Recipes)
4. Complete DMEM (see Recipes)
5. Reduced DMEM (see Recipes)
6. Differentiation medium with retinoic acid (see Recipes)
Recipes
1. Tris-HCl buffer (pH 8.5, 0.63 M)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Tris-HCl | 100 mg/mL (0.63 M) | 25.0 g |
| HCl | pH adjustment | ~4 mL (titrate to pH 8.5) |
| Deionized water | — | To 250 mL final volume |
Weigh 25.0 g of Tris-HCl (MW 157.6 g/mol) and transfer it to a clean 300 mL beaker containing ~200 mL of deionized water. Stir using a magnetic stirrer until completely dissolved. Slowly add concentrated HCl (~12 N) while monitoring the pH with a calibrated meter, adjusting to pH 8.5. As pH is temperature-dependent, record at the intended use temperature. Transfer the solution to a 250 mL volumetric flask and bring to volume with deionized water. Mix thoroughly. Filter-sterilize before final storage. Store at room temperature for up to 4 weeks or at 4 °C for up to 1 year.
2. PDA solution (0.15 mg/mL)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Dopamine hydrochloride | 0.15 mg/mL | 1.5 mg |
| Tris-HCl buffer (Recipe 1) | — | 10 mL |
Weigh 1.5 mg of dopamine hydrochloride and transfer it to a sterile Falcon tube containing 10 mL of Tris-HCl buffer to generate polydopamine (PDA) solution (Recipe 1). Stir the solution gently until all solids are completely dissolved. Prepare the PDA solution fresh before each experiment to avoid oxidation. Short-term storage at 4 °C is possible for up to 24 h. Handle under minimal light exposure to maintain stability. The recipe can be scaled up to facilitate more accurate weighing of dopamine hydrochloride.
3. RA stock solution (5 mM)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| All-trans retinoic acid | 5 mM | 50 mg |
| 95% ethanol | — | 33.3 mL |
Weigh 50 mg of RA powder and transfer it to a clean, amber bottle. Add 33.3 mL of 95% ethanol and gently swirl or invert until fully dissolved. RA is sensitive to light, heat, and air; handle under minimal light exposure and use a tightly sealed dark bottle to prevent degradation. Aliquot to prevent freeze-thaw cycles. Store at -20 °C for long-term storage. Prepare working dilutions freshly before experiments (see Recipe 6).
4. Complete DMEM
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| DMEM (+ 4.5 g/L D-Glucose) | 89% | 450 mL |
| FBS | 10% | 50 mL |
| Penicillin-streptomycin | 1% | 5 mL |
| Total | 100% | 505 mL |
Supplement 450 mL of DMEM with 50 mL of heat-inactivated FBS and 5 mL of penicillin-streptomycin. Mix gently to avoid foaming. Store at 2–8 °C for 2–4 weeks. Use sterile technique.
5. Reduced DMEM
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| DMEM (+ 4.5 g/L D-Glucose) | 98% | 450 mL |
| FBS | 1% | 5 mL |
| Penicillin-streptomycin | 1% | 5 mL |
| Total | 100% | 460 mL |
Supplement 450 mL of DMEM with 5 mL of heat-inactivated FBS and 5 mL of penicillin-streptomycin. Mix gently to avoid foaming. Store at 2–8 °C for 2–4 weeks. Use sterile technique.
6. Differentiation medium with retinoic acid
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Reduced DMEM medium (Recipe 5) | — | 5 mL |
| RA stock solution (Recipe 4) | 10 μM (500×) | 10 μL |
Add 10 μL of RA stock (5 mM in 95% ethanol) to 5 mL of prewarmed reduced DMEM (Recipe 5). Mix gently to ensure uniform distribution. Use immediately and protect from light. Prepare fresh before experiments and filter through a 0.22 μm membrane (optional) to ensure sterility.
Laboratory supplies
1. Falcon® 25 cm2 polystyrene tissue culture flasks, sterile, vent cap, canted neck (Corning, catalog number: 353108)
2. NuncTM 15 mL polypropylene centrifuge tubes, sterile, bulk bag (Thermo Scientific, catalog number: 339650)
3. Corning® 500 mL vacuum filter/storage bottle system, 0.22 μm pore, 33.2 cm2 CA membrane, sterile (Corning, catalog number: 430769)
4. Disposable syringe without needle, 10 mL (Air-Tite)
5. Sterile PES syringe filters, 0.22 μm (Fisher Scientific, catalog number: 09-720-511)
6. 55 mm glass-bottom dish with 30 mm micro-well, #1.5 cover glass (Cellvis, catalog number: D60-30-1.5-N)
7. Petri dishes, 90 mm × 15 mm (SPL)
Equipment
1. Class II biological safety cabinet (ESCO)
2. CO2 incubator (Thermo Scientific, model: 3110)
3. Vacuum oven (Thermo Scientific, model: 3608-5, 0.7 cu. ft.)
4. Voice coil actuator stage (Zaber, model: X-DMQ12L-AE55D12)
5. UV sterilizer or UV crosslinker
6. Analytical balance (METTLER TOLEDO, catalog number: 30823718)
7. Vortex mixer (Fisher Scientific, catalog number: 02215365)
8. Sterile forceps and scalpel
9. Biopsy punch (Ø 3 mm) (Qiagen, catalog number: WB100039)
10. Square punch (10 mm × 10 mm) (Walfront, catalog number: B0BRYB3BJK)
11. Water bath, 37 °C
12. Freezer, -20 °C
13. Refrigerator, 2–8 °C
14. Microscope capable of fluorescence microscopy (e.g., Nikon Ti2)
Software and datasets
1. ImageJ (National Institutes of Health, Version 1.54f); free, open-source image analysis software; no license required
2. MATLAB (MathWorks, Version R2023b); requires a paid license (academic, individual, or institutional). Free alternatives include GNU Octave (open-source)
3. All data and code have been deposited to GitHub: https://github.com/Carleton-CTE-Lab/Carleton-Cell-Stretcher (11/8/2025)
Procedure
文章信息
稿件历史记录
提交日期: Nov 9, 2025
接收日期: Dec 18, 2025
在线发布日期: Jan 7, 2026
出版日期: Feb 5, 2026
版权信息
© 2026 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Kang, G., Delgado, D., Petel, O. E. and Harris, A. R. (2026). Quantifying Mechanical Strain–Induced Membrane Damage in Early Neuronal Cells Using an In Vitro Traumatic Brain Injury Model. Bio-protocol 16(3): e5580. DOI: 10.21769/BioProtoc.5580.
分类
力学生物学
神经科学 > 神经系统疾病 > 动物模型
细胞生物学 > 组织分析 > 损伤模型
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










