- EN - English
- CN - 中文
Optimized Method for High-Quality Isolation of Single-Nuclei From Mosquito Fat Body for RNA Sequencing
一种从蚊子脂肪体中高质量分离单细胞核用于RNA测序的优化方法
(§Technical contact: stephanie.serafimdecarvalho@nih.gov) 发布: 2026年01月05日第16卷第1期 DOI: 10.21769/BioProtoc.5564 浏览次数: 260
评审: Anonymous reviewer(s)

相关实验方案

用Tagmentation Assisted Fragmentation法从K562细胞和果蝇神经母细胞的有限起始材料中提取芯片序列
Junaid Akhtar [...] Steffen Albrecht
2020年02月20日 4732 阅读
Abstract
Single-cell and single-nucleus RNA sequencing are revolutionizing our understanding of cellular biology. The identification of molecular markers, single-cell transcriptomic profiling, and differential gene expression at the cellular level has revealed key functional differences between cells within the same tissue. However, tissue dissociation remains challenging for non-model organisms and for tissues with unique biochemical properties. For example, the mosquito fat body, which serves functions analogous to mammalian adipose and liver tissues, consists of trophocytes—large, adipocyte-like cells whose cytoplasm is filled with lipid droplets. Conventional enzymatic dissociation methods are often too harsh for these fragile cells, and their high lipid content can interfere with reagents required for single-cell transcriptomic analysis. Single-nucleus RNA sequencing (snRNA-seq) offers an alternative strategy when intact cells with high-quality RNA cannot be obtained by enzymatic or mechanical dissociation. Here, we present an optimized reproducible methodology for nuclei isolation from the fat body of Anopheles gambiae mosquitoes, enabling high-quality snRNA-seq. Our approach involves tissue fixation and lipid removal, followed by cell lysis and nuclei purification using a sucrose cushion. We validated this protocol on both sugar-fed and blood-fed samples, established quality metrics to remove potential ambient RNA contamination, and demonstrated that snRNA-seq using this method yields high-quality sequencing results.
Key features
• Optimized nuclei isolation using methanol fixation and lipid removal enables efficient nuclei extraction from the fragile, lipid-rich fat body tissue of Anopheles gambiae.
• We validated this procedure in sugar-fed and blood-fed samples, yielding high-quality single-nucleus RNA sequencing data with high gene counts and low mitochondrial RNA content.
• Robust quality metrics allow effective filtering of ambient RNA, enhancing transcriptomic accuracy across different physiological states.
Keywords: Mosquito (蚊子)Graphical overview
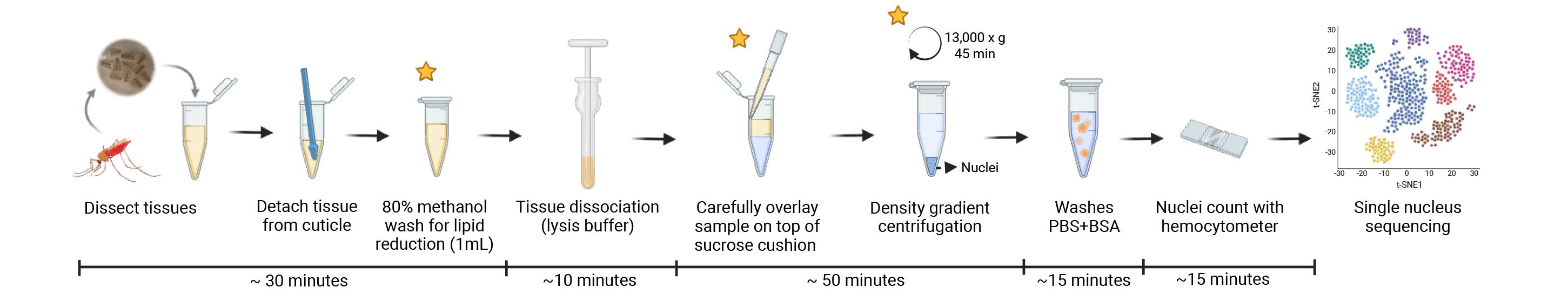
Tissue dissociation and nucleus isolation from mosquito fat body and associated tissues for single-nucleus RNA sequencing applications. Stars represent the critical steps of the protocol.
Background
Single-cell RNA sequencing (scRNA-seq) has transformed our understanding of cellular heterogeneity by enabling high-resolution molecular profiling at the level of individual cells. While conventional bulk RNA sequencing offers deep insights into transcription regulation, it only captures the average gene expression across a population of cells, masking potential cell-to-cell variability. In contrast, single-cell omics allow for the analysis of transcriptomes from individual cells, generating distinct gene expression profiles that reveal cell-specific functions and dynamic states within complex tissues.
Mosquitoes are the main vectors of arboviruses and parasites, impacting millions of people each year. Blood feeding is a critical behavior in female mosquitoes, as it provides the nutrients necessary for egg development; however, it also enables the transmission of pathogens when mosquitoes feed on infected hosts. The ingestion of a blood meal triggers profound physiological changes, particularly in the fat body, a central metabolic and reproductive tissue in mosquitoes [1–3]. Functionally similar to the liver and adipose tissue in mammals, the fat body rapidly shifts its gene expression after a blood meal, including the production of yolk proteins like vitellogenin that are essential for reproduction. Trophocytes—large cells with the cytoplasm filled with extensive lipid droplets—constitute the main cell type of the fat body [1–4]. These are adipocyte-like cells with high lipid content and a fragile structure, which makes dissociation and single-cell isolation very difficult, leading to poor cell recovery, low viability, and reduced RNA integrity when using standard single-cell isolation procedures.
A critical step in scRNA-seq is an efficient dissociation of tissues into a high-quality single-cell suspension. However, certain tissue types—such as adipose tissue—pose a significant technical challenge due to their large size (20–300 μm) [5], structural fragility, and high lipid content [6]. Enzymatic dissociation methods (e.g., collagenase or trypsin digestion) are often used to dissociate tissues, but they can introduce technical artifacts, damage fragile cells, and reduce RNA integrity, especially in lipid-rich tissues [6–8]. These limitations may result in poor cell viability and biased transcriptomic profiles, making conventional scRNA-seq challenging for these tissues. Single-nucleus RNA sequencing (snRNA-seq) offers several advantages for challenging tissues, including those that are fragile, lipid-rich, or difficult to dissociate. snRNA-seq avoids dissociation-induced transcriptional artifacts, preserves representation of large or fragile cell types, and is compatible with frozen or archived tissues [9,10]. However, because nuclear RNA is enriched for pre-mRNAs and lacks some cytoplasmic transcripts, snRNA-seq typically detects fewer genes per nucleus and may underestimate transcripts with strong cytoplasmic localization [11,12]. Thus, while snRNA-seq provides a powerful alternative when whole-cell isolation is not feasible, it also requires accounting for nuclear-specific transcriptional features during data analysis.
Prior studies have explored single-nucleus transcriptomic approaches in insects. In Aedes aegypti, single-nucleus RNA sequencing has been applied to both brain and midgut tissues, demonstrating the feasibility of nuclear profiling in mosquitoes [13,14]. However, quality metrics such as nuclei yield, ambient RNA contamination, and percentage of mitochondrial transcripts are poorly reported. Additionally, snRNA-seq has been performed in the fruit fly Drosophila melanogaster and the Bombyx mori moth fat body tissues, profiling cell-type diversity and functional heterogeneity [15,16]. An optimized Drosophila nuclei isolation protocol reported the recovery of ~106 nuclei from 40 fat body tissues, with improved levels of mitochondrial contamination (<20%) and unique transcripts (>200) compared to enzymatic methods [17]. Despite these advances, isolating intact cells from the mosquito fat body remains technically challenging. To date, no standardized or optimized nuclei isolation protocol has been developed for Anopheles gambiae fat body. Here, we present a reproducible nuclei-based method that yields high-quality, intact nuclei from Anopheles gambiae fat body with consistent recovery across samples. Our protocol recovers approximately 1 × 106 nuclei from 25 body wall tissues, maintains nuclear membrane integrity, and supports low levels of ambient RNA contamination, enabling robust snRNA-seq profiling. In addition, the workflow is compatible with both fresh and methanol-stabilized tissues and is completed rapidly without enzymatic digestion. These features collectively provide a practical and reliable approach for high-quality transcriptomic analysis of this metabolically and immunologically important tissue.
Materials and reagents
Biological materials
1. Anopheles gambiae (G3, CDC strain) dissected body walls (25 tissues) from adult females
Reagents
1. Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: C1016-500G)
Caution: Irritant to the eyes.
2. 2 M potassium chloride (KCl) (Thermo Fisher Scientific, catalog number: AM9640G, 100 mL)
3. 1 M HEPES, pH 7.4 (Teknova, catalog number: H1030, 1,000 mL)
4. Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266-100G)
5. 5 M sodium chloride (NaCl) (KD Medical, catalog number: RGF-3270, 1,000 mL)
6. Sodium bicarbonate (NAHCO3) (Sigma-Aldrich, catalog number: S6297-250G)
7. Sodium phosphate monobasic (NaH2PO4) (Sigma-Aldrich, catalog number: S3139-250G)
8. Sucrose (Sigma-Aldrich, catalog number: S0389-500G)
9. D-(+)-Trehalose dihydrate (Sigma-Aldrich, catalog number: T0167-25G)
10. Methanol (Thermo Fisher Scientific, catalog number: A412P-4, 4 L)
Caution: Flammable liquid and vapor. Toxic by inhalation, in contact with skin, and if swallowed. Handle inside of a safety cabinet.
11. Nuclease-free water (NFW), no DEPC (Thermo Fisher Scientific, catalog number: AM9938)
12. Nuclei Isolation kit, Nuclei PURE Prep (Sigma-Aldrich, catalog number: NUC201-1KT)
13. Dithiothreitol (DTT) (Thermo Fisher Scientific, catalog number: D1532, 1 g)
Caution: Irritant to skin and eyes. Harmful if swallowed.
14. Phosphate buffer saline (PBS) 10× (90 g NaCl, 1.44 g KH2PO4, 7.95 g NaHPO4 in 1 L of water), pH 7.4 (KD-Medical, catalog number: RGF-3210)
15. RNASE inhibitor, protector RNASE inhibitor 10,000 units (Sigma-Aldrich, catalog number: 3335402001)
16. Ultrapure BSA 50 mg/mL (Thermo Fisher Scientific, catalog number: AM2618)
17. RNase Away (Molecular Bioproducts, catalog number: 7000)
18. Trypan blue stain, 0.4% (Gibco, catalog number: 15250061, 100 mL)
19. Triton X-100, molecular biology (Millipore, catalog number: 648466, 50 mL)
Solutions
1. Hemolymph-like saline (HLS buffer) (see Recipes)
2. 80% methanol solution (see Recipes)
3. Lysis buffer (see Recipes)
4. 1.8 M sucrose cushion solution (see Recipes)
5. Washing buffer 1× (see Recipes)
6. Resuspension buffer 1× (see Recipes)
Recipes
Note: Keep all solutions on ice at all times to maintain RNA integrity during the procedure. Prepare solutions fresh before nucleus isolation. Storage is not recommended, except for the HLS buffer.
1. HLS buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| CaCl2 0.5 M | 2 mM | 0.4 mL |
| KCl 2 M | 5 mM | 0.25 mL |
| HEPES 1 M, pH 7.4 | 5 mM | 0.5 mL |
| MgCl2 0.5 M | 8.2 mM | 1.64 mL |
| NaCl 5 M | 108 mM | 2.16 mL |
| NaHCO3 0.5 M | 4 mM | 0.8 mL |
| NaH2PO4 0.5 M | 1 mM | 0.2 mL |
| Sucrose 0.5 M | 10 mM | 2 mL |
| Trehalose 1 M | 5 mM | 0.5 mL |
| Nuclease-free water | n/a | 91.55 mL |
| Total | n/a | 100 mL |
Notes:
1. Adjust the pH to 7.5, filter, and store aliquots at 4 °C.
2. Use solutions of CaCl2 and MgCl2 instead of powders to avoid precipitation in the saline.
3. This buffer was previously proposed by Singleton and Woodruff [18] to mimic Drosophila hemolymph osmolarity.
2. 80% methanol solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Methanol | 80% | 8 mL |
| HLS buffer | n/a | 2 mL |
| Total | n/a | 10 mL |
Notes:
1. Each sample requires the use of 1 mL of 80% methanol solution.
2. This solution, used on step C1, is important to remove excess lipids.
Caution: Prepare and handle this solution inside a safety cabinet.
3. Lysis buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Nuclei PURE lysis buffer (from Nuclei PURE Prep kit) | n/a | 989 μL |
| DTT 1 M | 1 mM | 1 μL |
| Triton X-100 10% | 0.1% | 10 μL |
| Total | n/a | 1 mL |
Note: Each sample requires the use of 0.5 mL of lysis buffer.
4. 1.8 M sucrose cushion solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Nuclei PURE 2 M Sucrose cushion solution (from Nuclei PURE Prep kit) | 1.8 M | 5.4 mL |
| Nuclei PURE sucrose cushion buffer (from Nuclei PURE Prep kit) | n/a | 0.6 mL |
| Total | n/a | 6 mL |
Notes:
1. Each sample requires the use of 2.8 mL of 1.8 M sucrose cushion solution.
2. Mix with a pipette or vortex until the solution looks clear and homogeneous. This solution is viscous due to the high sucrose concentration.
5. Washing buffer 1×
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| PBS | 1× (150 mM, 1 mM KH2PO4, 5.5 mM NaHPO4) | 100 μL |
| Ultrapure BSA (50 mg/mL) | 10 mg/mL or 1% | 200 μL |
| RNase inhibitor (40 U/μL) | 0.2 U/μL | 5 μL |
| Nuclease-free water | n/a | 695 μL |
| Total | n/a | 1 mL |
Note: Each sample requires the use of 2 mL of washing buffer.
6. Resuspension buffer 1×
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| PBS | 1× (150 mM, 1 mM KH2PO4, 5.5 mM NaHPO4) | 100 μL |
| Ultrapure BSA (50 mg/mL) | 2 mg/mL or 0.2% | 40 μL |
| RNase inhibitor (40 U/μL) | 0.2 U/μL | 5 μL |
| Nuclease-free water | n/a | 855 μL |
| Total | n/a | 1 mL |
Laboratory supplies
1. 1.5 mL microtubes (Axygen, catalog number: MCT-150-C-S)
2. 10 μL micropipette tips (TipOne 10 μL, graduated filter tips) (USA Scientific, catalog number: 1180-3810)
3. 200 μL micropipette tips (TipOne 200 μL graduated filter tips) (USA Scientific, catalog number: 1180-8810)
4. 1,250 μL micropipette tips (Purepoint barrier tips FT1250) (Alkali Scientific, catalog number: FT1250)
5. C-Chip disposable hemocytometer (iNCYTO, catalog number: DHC-N01-5 NI, Neubauer Improved)
6. Laboratory pipettes (10, 20, 200, and 1,000 μL)
7. Transfer pipette, sterile, polyethylene (Millipore Sigma, catalog number: Z350818-500EA)
8. WHEATON Dounce tissue grinder (WHEATON, catalog number: 357538, 1 mL)
9. Pellet pestles (Fisher Scientific, catalog number: 12-141-363)
10. 40 μm filter (Pluriselect, catalog number: 43-10040-40)
11. 20 μm filter (Pluriselect, catalog number: 43-10020-40)
Equipment
1. Refrigerated centrifuge (Eppendorf, model: 5417 R)
2. Rotor (Eppendorf, catalog number: F45-30-11)
3. Transmitted light and epifluorescence microscopes
Software and datasets
1. Cell ranger (10X genomics, version 7.0.0)
2. Seurat R package (Satija Lab, version 4.2.1)
3. DecontX R package (Campbell Lab, version 1.6.0)
4. All data have been deposited to the SRA database under BioProject accession number PRJNA1288431 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA1288431?reviewer=he84ttfsjqcmttfp3ccg035vi8)
Procedure
文章信息
稿件历史记录
提交日期: Oct 27, 2025
接收日期: Dec 7, 2025
在线发布日期: Dec 16, 2025
出版日期: Jan 5, 2026
版权信息
© 2026 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
de Carvalho, S. S., McNinch, C. and Barillas-Mury, C. (2026). Optimized Method for High-Quality Isolation of Single-Nuclei From Mosquito Fat Body for RNA Sequencing. Bio-protocol 16(1): e5564. DOI: 10.21769/BioProtoc.5564.
分类
分子生物学 > RNA > RNA 测序
细胞生物学 > 细胞器分离 > 细胞核
细胞生物学 > 单细胞分析
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










