- EN - English
- CN - 中文
Reproducible Sample Preparation of Virus-Infected Cells for Cryo-FIB/ET Using Manual Plunge Freezing
利用手动投入冷冻法对病毒感染细胞进行可重复制备以用于冷冻聚焦离子束/电子断层扫描
(§Technical contact: nathalie.lavoie@tufts.edu) 发布: 2026年01月05日第16卷第1期 DOI: 10.21769/BioProtoc.5563 浏览次数: 329
评审: Abhilash PadavannilAnonymous reviewer(s)

相关实验方案

TZA, 一种基于敏感报告因子细胞学检测方法, 可用于准确快速量化静息CD4+ T细胞中可诱导的且复制能力强的潜伏HV-1
Anwesha Sanyal [...] Phalguni Gupta
2019年05月20日 7794 阅读
Abstract
Most viruses extensively remodel their host cells to establish productive infection. Visualization of virus-induced cellular remodeling by electron microscopy (EM) has been revolutionized in recent years by advances in cryo-focused ion beam (cryo-FIB) milling paired with cryo-electron tomography (cryo-ET). As cryo-FIB/ET becomes more widely available, there is a need for beginner-friendly guides to optimize the preparation of virus-infected mammalian cells on EM grids. Here, we provide an in-house protocol for new users for preparing samples of cells infected with herpes simplex virus 1 (HSV-1) for cryo-FIB/ET. This protocol guides users in how to seed infected cells onto grids, blot, and plunge-freeze grids using basic, manual equipment. It also provides tips on how to screen and prioritize grids for efficient milling and data collection.
Key features
• A beginner-friendly protocol for users without access to a cryo-EM core/suite at their institution that utilizes basic equipment.
• This protocol focuses on optimizing cell seeding and blotting to yield grids with thin ice and evenly distributed cells.
• Grids prepared using this protocol can be used for focused ion beam milling.
Keywords: Cryo-ET (cryo-electron tomography) (冷冻电子断层扫描技术)Graphical overview
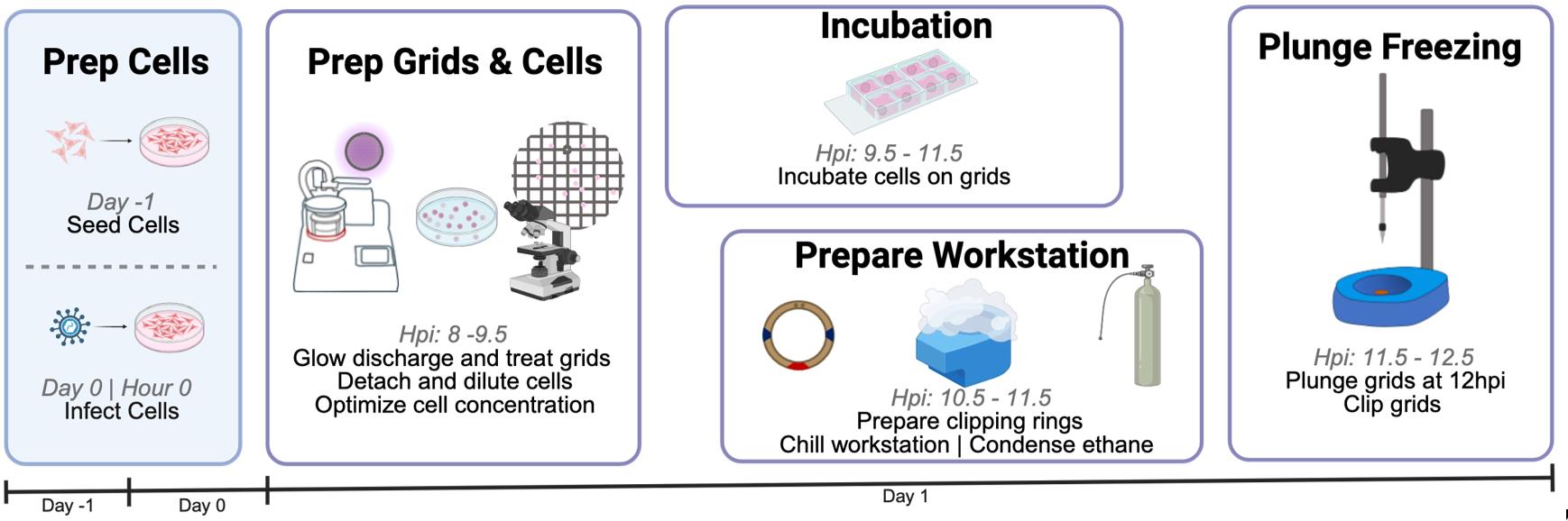
Schematic overview of sample and grid preparation for cryo-electron tomography (cryo-ET). This protocol provides detailed guidance on each critical step of preparation to increase the success of grid generation for plunge freezing. It requires three days from cell seeding to grid clipping and storage. Eight hours post-infection (hpi), users begin preparation for plunge freezing by transferring infected cells onto grids. From 9.5 to 11.5 hpi, users incubate cells on grids and set up the plunge freezer. In this protocol, cells are frozen at 12 hpi and then clipped.
Background
Cryo-focused ion beam (cryo-FIB) milling has dramatically expanded the targets accessible to cryo-electron tomography (cryo-ET) by enabling high-resolution imaging of the cellular interior. In particular, cryo-ET/FIB enables examination of virus-infected cells at an unprecedented level of detail [1–4]. Before the implementation of cryo-FIB, cryo-ET was largely limited to imaging peripheral regions of the cell. Access to the cellular interior, with limited sample distortion, permits researchers to determine the structures of key proteins in their native environment and provides insight into their cellular context. This access is crucial for studying viral replication and assembly in important cellular compartments, such as the nucleus. In the past five years, automation of FIB milling and data collection has increased the efficiency of this pipeline [5–7]. At the same time, software for cryo-ET data processing and analysis, such as RELION 5.0 [8] and AreTomo2 [9,10], has been introduced. Despite these advances on the processing end of the cryo-ET/FIB pipeline, sample preparation remains a major roadblock, especially for beginners. Sample preparation prior to cryo-FIB milling includes (1) seeding cells onto an EM grid and (2) freezing and clipping grids.
Success of cryo-ET requires thin, <200 nm lamellae produced by cryo-FIB milling. In turn, successful FIB milling requires a sample with optimal cell distribution on grids. Cell aggregation or excess buffer can increase sample thickness, requiring more time and a higher ion beam current on the dual-focused ion beam–scanning electron microscope (FIB-SEM), which can damage the cell. Therefore, it is critical to optimize cell seeding and grid blotting to maximize efficient use of dual-beam microscope time while ensuring cell integrity. This protocol focuses on the first two steps of sample preparation to prepare high-quality grids reproducibly for FIB-SEM.
Despite the existence of automated plunge freezers, such as Leica GP2 and ThermoFisher VitroBot, they may not be readily accessible to all interested users. Besides, manual plunge freezers offer some practical benefits. For instance, they are more portable, which is advantageous for users who work with BSL2-level samples and must plunge-freeze them inside a biosafety hood. In addition, they are more cost-effective, being typically 1/10th the price of automatic plunge freezers, making them accessible to more labs. Finally, manual plunge-freezing can be beneficial when working with fragile cells. However, manual plungers require more careful handling than automatic plungers to achieve high reproducibility. Poor handling can lower the number of grids suitable for milling.
This protocol was developed to prepare samples of mammalian cells infected with herpes simplex virus 1 (HSV-1) for cryo-FIB/ET to study a process termed “nuclear egress” that is unique to herpesviruses [11]. However, it can be readily adapted for use with uninfected mammalian cells after any treatment, such as chemical inhibitors. This protocol guides beginner-level users in how to seed infected cells onto grids, blot, and plunge-freeze grids using basic, manual equipment under BSL2 conditions. It emphasizes optimizations and provides troubleshooting suggestions for sample preparation. In our hands, the use of this protocol has drastically improved the yield of millable grids for cryo-ET/FIB. Published protocols that describe the use of a manual plunge freezer [12–15], handling liquid ethane [16], and using a clipping station [17] are important companions. Finally, protocols that provide guidance on growing cells on grids can be used in conjunction with this protocol [18–20].
Materials and reagents
Biological materials
1. Vero cells (ATCC, catalog number: CCL-81) or HeLa Cells (ATCC, catalog number: CCL-2)
Note: This protocol has been tested with both Vero and HeLa cells, but it could be applied to other adherent cells.
2. Herpes simplex virus 1 tdTomato-NLS (strain HSV-1 F-GS3217, which encodes a tdTomato-NLS fluorescent reporter under the control of a CMV promoter) (gift from Dr. Gregory Smith, Northwestern University)
Note: A strain encoding a fluorescent reporter enables the use of integrated fluorescence light microscopy (iFLM). Any HSV-1 or HSV-2 strain encoding a fluorescent reporter can be used instead of the one listed here. A strain lacking a fluorescent reporter can also be used, but without iFLM.
Reagents
1. Dulbecco’s modified Eagle’s medium (DMEM) (Cytiva, catalog number: SH30285.01); store at 4 °C
2. Penicillin-streptomycin solution, 100× (Cytiva, catalog number: SV30010); store at 4 °C
3. Fetal bovine serum (FBS), heat-inactivated (BioWest, catalog number: S1680-500); store at -20 °C
4. L-glutamine, 200 mM (Corning, catalog number: 25-005-CI); store at 4 °C
5. Phosphate-buffered saline (PBS), 1× (Cytiva, catalog number: SH30256.FS); store at room temperature
6. Trypsin, 0.05% protease solution (Cytiva, catalog number: SH30236.01); store at 4 °C
7. Trypan blue solution (Cytiva, catalog number: SV30084.01); store at room temperature
8. Poly-l-lysine solution, 0.1% (Sigma-Aldrich, CAS number: 25988-63-0); store at room temperature
9. Bleach (Clorox, catalog number: 30966); store at room temperature
Solutions
1. Complete DMEM (cell growth media) (see Recipes)
2. 20% (v/v) bleach (see Recipes)
3. 0.01% (v/v) poly-l-lysine (see Recipes)
Recipes
1. Complete DMEM (cell growth media)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| DMEM | 1× | 440 mL |
| FBS | 10% | 50 mL |
| Penicillin-streptomycin solution, 100× | 1× | 5 mL |
| L-glutamine, 200 mM | 2 mM | 5 mL |
| Total | 500 mL |
Remove a 60 mL aliquot of DMEM from the 500 mL bottle before adding any components, save it, and label it as “serum-free DMEM.” Store complete DMEM at 4 °C for up to 3 months.
2. 20% (v/v) bleach
Prepare fresh. For 200 mL, mix 40 mL of undiluted bleach with 160 mL of water. Store in a plastic bottle.
3. 0.01% (v/v) poly-l-lysine
Prepare fresh. For 200 μL, mix 20 μL of 0.1% poly-l-lysine with 180 μL of filtered, deionized water.
Laboratory supplies
1. 35 mm × 10 mm polystyrene tissue culture–treated dishes (CELLTREAT, catalog number: 229635)
2. 40 μm cell strainers (NEST Scientific, catalog number: 258369)
3. 50 mL centrifuge tubes (CELLTREAT, catalog number: 229421)
4. 15 mL centrifuge tubes (CELLTREAT, catalog number: 229411)
5. 1.5 mL microcentrifuge tubes (Bio Plas, catalog number: 4030)
6. 150 mm × 20 mm polystyrene tissue culture–treated dishes (CELLTREAT, catalog number: 229651)
7. UltrAuFoil, R 2/2; Gold 200 mesh grids (Quantifoil, catalog number: Q250AR2A)
8. Hybrid grid box (NanoSoft, catalog number: 14021008)
9. Fisherbrand qualitative grade filter paper, P4 (FisherScientific, catalog number: 09-803-6D)
10. 8-well clear chamber slides (NEST Scientific, catalog number: 230108)
11. Dumont tweezers style N5 (Electron Microscopy Sciences, catalog number: 0203-N5-PO)
12. Spearlab Foam Dewar, standard vessel (MiTeGen, SKU: M-FD-800)
13. UN1977 liquid nitrogen (MedTech)
14. Ethane, research grade (AirGas, catalog number: ET RP35)
15. Brass gas regulator (AirGas, catalog number: Y12244A350-AG)
16. VWR® CryoGuard cryogenic gloves (VWR, catalog number: 97008-216)
17. Deluxe surgical mask (Uline, catalog number: S-22137)
18. Colored labeling tape (Fisher Scientific, catalog number: 15-901-R)
19. Heavy duct tape (Amazon, ASIN: B07WHV2F9C)
20. Autogrid tweezers (NanoSoft, catalog number: 23021002)
21. Pin lid manipulator tool (NanoSoft, catalog number: 27011001)
22. C-clips (NanoSoft, SKU: 11011002)
23. Cryo-FIB Autogrid Rings (Thermo Fisher Scientific, SKU: 1205101)
Note: Must be purchased directly from a Thermo Fisher representative; alternatives are available from Ted Pella/NanoSoft, but these should be discussed with EM core managers.
24. Colored cryo-markers, ethanol resistant (Electron Microscopy Sciences, catalog number: 62050)
25. Screwdriver for puck lid lock (MiTeGen, SKU: M-CEMG2-LLSD)
Equipment
1. Biosafety cabinet (Nuaire, model: NU-425-400)
2. CO2 cell culture incubator (Thermo Scientific, model: Heracell VIOS 160i)
3. Fluorescent microscope (Leica, model: DM IL LED)
4. Hausser Scientific phase hemacytometer (Fisher Scientific, catalog number: 02-671-10)
5. ZEROSTAT3 anti-static gun (Ted Pella, catalog number: 54610)
6. PELCO easiGlowTM glow-discharge system for cryo-EM (Ted Pella, catalog number: 91000S)
7. Liquid dewar (4 L) (MidSci, catalog number: Q250AR2A)
8. Manual plunge freezer, foot-pedal operation (Neptune Fluid Flow Systems, catalog number: MP1)
9. Vitrification dewar including metal parts (NanoSoft, catalog number: 21021006)
10. 10× magnifying glass with light and clamp (TOMSOO, Amazon, ASIN: B0B8RZWW8X)
11. Clipping station (NanoSoft, catalog number: 25011001)
12. 4-way flipper racks (ThermoFisher Scientific, catalog number: 8850)
13. Hair dryer (Amazon, ASIN: B00GUAEERO)
14. MiTeGen Cryo-EM Puck, generation 2.0 (MiTeGen, SKU: M-CEMG2-1)
Note: SubAngstrom also supplies cryo-EM pucks with distinct racks from MiTeGen. Contact your EM core to determine their storage preference.
15. Transport cane for Cyro-EM puck (MiTeGen, SKU: M-CEMTPC-1)
16. High-capacity (HC) liquid nitrogen refrigerator (MiTeGen, SKU: TW-HC34)
17. Aquilos Cryo-FIB (ThermoFisher)
Software and datasets
1. ThermoFisher xT (Beam control)
2. ThermoFisher Maps 3.21
Procedure
文章信息
稿件历史记录
提交日期: Oct 9, 2025
接收日期: Dec 1, 2025
在线发布日期: Dec 16, 2025
出版日期: Jan 5, 2026
版权信息
© 2026 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Lavoie, N. R. and Heldwein, E. E. (2026). Reproducible Sample Preparation of Virus-Infected Cells for Cryo-FIB/ET Using Manual Plunge Freezing. Bio-protocol 16(1): e5563. DOI: 10.21769/BioProtoc.5563.
分类
微生物学 > 微生物细胞生物学 > 基于细胞的分析方法
细胞生物学 > 细胞成像
生物物理学 > 电子冷冻断层扫描
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










