- EN - English
- CN - 中文
Creating Loss-of-Function Mutants of Neurospora crassa Using a Novel CRISPR/Cas9 System
利用新型CRISPR/Cas9系统构建粗糙脉孢菌的功能缺失突变体
发布: 2026年01月05日第16卷第1期 DOI: 10.21769/BioProtoc.5562 浏览次数: 193
评审: Alba BlesaAnonymous reviewer(s)
Abstract
Since its introduction, the CRISPR/Cas9 system has been used in many organisms for precise and rapid genome editing, as well as for editing multiple genes at once. This targeted mutagenesis makes it easy to analyze the function of a gene of interest (goi). The standard method for genetic manipulation of the model organism Neurospora crassa has been homologous recombination. It is well established and widely used to create knock-out or overexpression mutants. The recently developed CRISPR/Cas9 system is an addition to the toolkit for genetically manipulating N. crassa. For this protocol, a strain stably expressing the Cas9 endonuclease is required. After designing the gRNA with the online tool CHOP-CHOP, a synthetic gRNA is used to transform macroconidia via electroporation. Combining the goi-gRNA with a gRNA targeting the csr-1 gene as a selection marker allows for easy identification of colonies with mutations at the target site of the goi, since the obtained resistance to Cyclosporin A (CsA) allows for selecting editing events. The mutation type can be detected by PCR of the edited gene region followed by Sanger sequencing. This system is fast and easy to handle, offering an attractive alternative to homologous recombination, especially for targeting multiple genes simultaneously.
Key features
• This protocol allows the use of CRISPR/Cas9 in Neurospora crassa to create loss-of-function mutants.
• It can be used to create loss-of-function mutants of multiple genes in one round of transformation.
• Time-saving mutagenesis without the need for vector construction.
• In combination with csr-1 as a selection marker, the screening for successfully edited genes of interest is reduced.
Keywords: CRISPR/Cas9 (CRISPR/Cas9)Graphical overview
Background
Neurospora crassa is a widely used eukaryotic model organism because of its ease of growth, genetic accessibility, and rapid life cycle. It is commonly employed in genetic research, where its genome is modified to study gene functions [1].
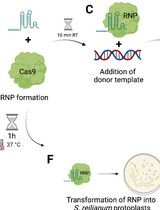
Traditionally, random mutagenesis methods such as UV exposure or chemicals have been used for genetic modification, which causes multiple unpredictable mutations. Identifying these mutations is time-consuming, and pinpointing the exact mutation responsible for a phenotype is difficult. Therefore, targeted mutagenesis is preferred. In N. crassa, homologous recombination (HR) has become the preferred method for creating targeted gene knockouts or overexpression lines. This involves transforming the organism with a vector containing homologous flanking sequences, the gene of interest, and/or a selection marker [2]. For higher efficiency, strains lacking nonhomologous end joining (NHEJ) are often used [3,4]. After transformation and the generation of homokaryotic transformants, validation is performed via PCR or Southern blot [3,5]. The whole process can take up to seven weeks, is labor-intensive, and is less efficient if NHEJ-deficient strains are not used [6]. The CRISPR/Cas9 system offers a faster and more precise alternative. It involves the Cas9 endonuclease and a guide RNA (gRNA) that directs editing to specific DNA sequences. When Cas9 induces a double-strand break, the cell repairs it either through NHEJ, which may introduce small insertions or deletions (indels), or homology-directed repair (HDR), which allows for the integration of new DNA sequences when a suitable donor DNA is available [7].
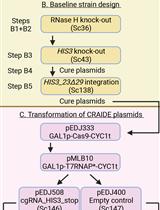
Although the CRISPR/Cas9 system has been adapted for various fungi [8], it has not yet been widely used in N. crassa. Another established system for N. crassa requires co-transforming multiple plasmids carrying Cas9, gRNA, and donor DNA, making it complex and labor-intensive [9]. In contrast, the system described here simplifies this process by integrating Cas9 directly into the genome and introducing naked synthetic gRNA via electroporation of macroconidia, eliminating the need to clone and co-transform Cas9/gRNA plasmids [10].
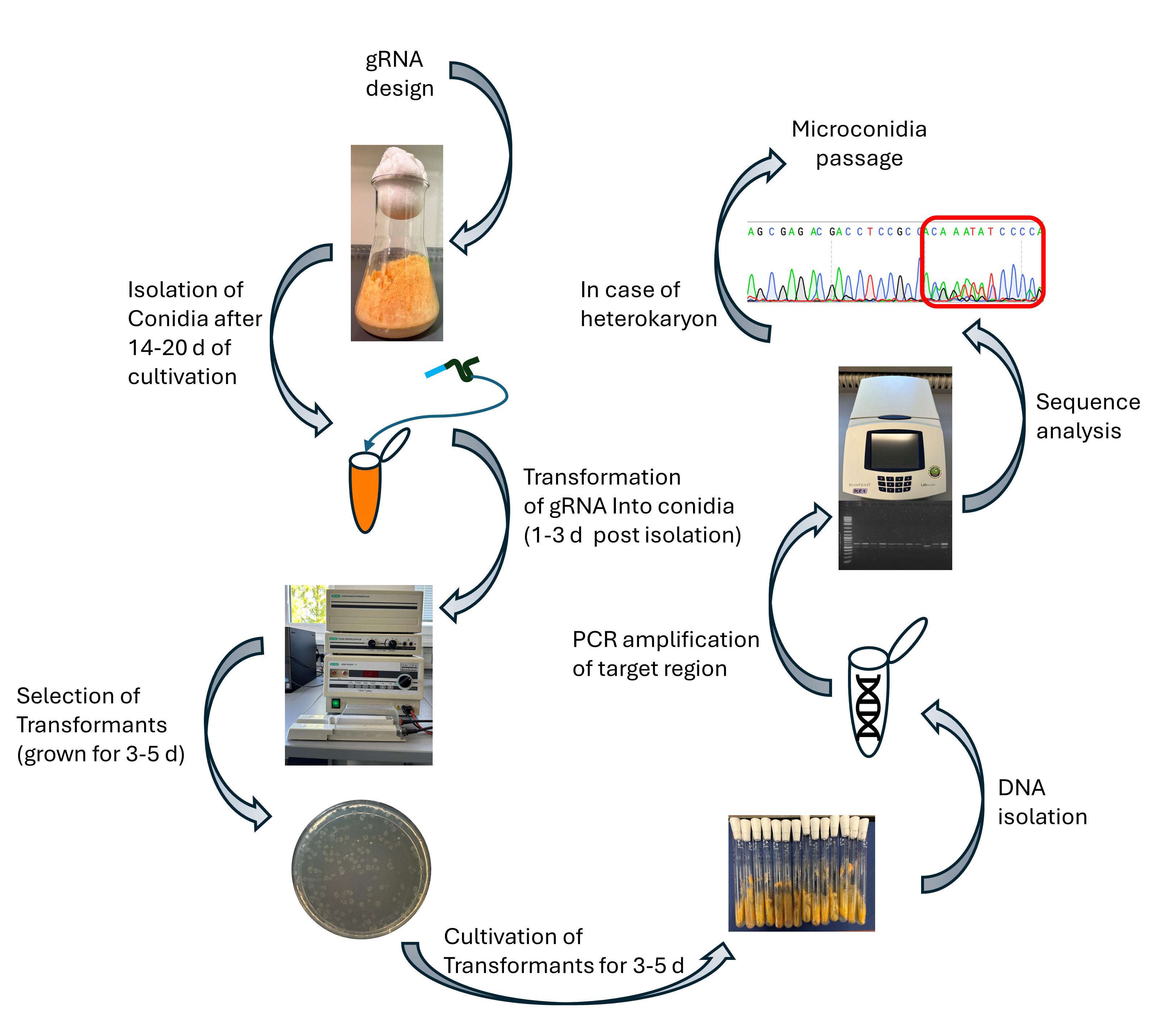
One concern with stable Cas9 expression is the potential for off-target mutations [11]. Studies in fungi suggest that limiting the duration of Cas9-gRNA complex presence can reduce this risk [12,13]. Even if Cas9 remains in the cells, controlling gRNA expression helps minimize off-target effects. To further reduce potential off-target mutations after successful mutagenesis, outcrossing the Cas9 sequence may be advisable. Co-transformation of multiple gRNAs enables simultaneous editing events at once. This ability allows the use of the csr-1 gene as a selection marker in N. crassa. It involves co-transforming a gRNA targeting csr-1 with a gRNA targeting the gene of interest (goi). If csr-1 is successfully edited and becomes non-functional, the fungus exhibits resistance to Cyclosporin A (CsA). This resistance allows selection for edited strains using CsA-containing media, simplifying the identification process. Additionally, mutations in csr-1 typically produce homokaryons [14]. Traditionally, isolating homokaryotic strains involves time-consuming steps such as crossing, microconidia isolation, or serial transfers of macroconidia. However, these steps can be bypassed by using the CRISPR/Cas9 system with csr-1 as a marker. Overall, this approach offers a time- and effort-saving, user-friendly method for generating loss-of-function mutants in N. crassa.
Materials and reagents
Biological materials
1. Neurospora crassa strain NcCas9SG [available at the Fungal Genetics Stock Center (FGSC); reference #27376]
Reagents
1. Sucrose (Roth, CAS number: 57-50-1)
2. Agar-agar (Roth, CAS number: 9002-18-0)
3. Citric acid (Roth, CAS number: 5949-29-1)
4. ZnSO4·7H2O (Merck, CAS number: 7446-20-0)
5. Fe(NH4)2(SO4)2·6H2O (Merck, CAS number: 7783-85-9)
6. CuSO4·5H2O (Merck, CAS number: 7758-99-8)
7. MnSO4·H2O (Roth, CAS number: 10034-96-5)
8. H3BO3 (Roth, CAS number: 10043-35-3)
9. Na2MoO4·2H2O (Merck, CAS number: 10102-40-6)
10. Chloroform (Roth, CAS number: 67-66-3)
11. Na3-Citrate·2H2O (Roth, CAS number: 6132-04-3)
12. KH2PO4 (Roth, CAS number: 7778-77-0)
13. NH4NO3 (Roth, CAS number: 6484-52-2)
14. MgSO4·7H2O (Merck, CAS number: 10034-99-8)
15. CaCl2·2H2O (Roth, CAS number: 10035-04-8)
16. Biotin (Roth, CAS number: 58-85-5)
17. TRIS (Roth, CAS number: 77-86-1)
18. NaCl (Roth, CAS number: 7647-14-5)
19. KCl (Roth, CAS number: 7447-40-7)
20. EtOH p.a. (Roth, CAS number: 64-15-7)
21. EDTA (Roth, CAS number: 6381-92-6)
22. Na-iodoacetate (Merck, CAS number: 305-53-3)
23. KNO3 (Roth, CAS number: 7757-79-1)
24. K2HPO4 (Roth, CAS number: 7758-11-4)
25. KOH (Roth, CAS number: 1310-58-3)
26. Cyclosporin A (biomol, catalog number: AG-CN2-0079-M100)
27. Sorbitol (Roth, CAS number: 50-70-4)
28. Tween 80 (Roth, CAS number: 9005-65-6)
29. Alt-RTM CRISPR-Cas9 tracrRNA (20 nmol) (IDT, catalog number: 1072533)
30. Duplex buffer (IDT, catalog number: 11-01-03-01)
Solutions
1. 50× Vogel’s solution (see Recipes)
2. Trace elements solution (TES) (see Recipes)
3. Vogel’s minimal medium + sucrose (VMM+S) (see Recipes)
4. Vogel’s minimal medium + sorbose-fructose-glucose + cyclosporin A (VMM+SGF+CsA) (see Recipes)
5. Sorbose-fructose-glucose solution (SGF) (see Recipes)
6. Top agar (see Recipes)
7. TPS buffer (see Recipes)
8. MKM medium (see Recipes)
9. 2× Synthetic crossing (SC) solution (see Recipes)
11. 1 M sorbitol + 0.25% Tween 80 (see Recipes)
Recipes
1. 50× Vogel’s solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Na3-Citrate·2H2O | 510 mM | 75 g |
| KH2PO4 | 1.84 M | 125 g |
| NH4NO3 | 1.25 M | 50 g |
| MgSO4·7H2O | 40.6 mM | 5 g |
| CaCl2·2H2O* | 34 mM | 2.5 g |
| TES | 1% (v/v) | 5 mL |
| Biotin | 0.1 mM | 12.5 mg |
| Chloroform** | 0.2% (v/v) | 1 mL |
| Autoclaved ddH2O | n/a | Add up to 500 mL |
| Total | n/a | 500 mL |
*Before adding, dissolve in 50 mL of ddH2O.
**Chloroform is added as a preservative and is optional.
Store Vogel’s solution in the dark at room temperature (RT).
2. Trace element solution (TES)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Citric acid | 5% (w/v) | 5 g |
| ZnSO4·7H2O | 5% (w/v) | 5 g |
| Fe(NH4)2(SO4)2·6H2O | 1% (w/v) | 1 g |
| CuSO4·5H2O | 0.25% (w/v) | 250 mg |
| MnSO4·H2O | 0.05% (w/v) | 50 mg |
| H3BO3 | 0.05% (w/v) | 50 mg |
| Na2MoO4·2H2O | 0.05% (w/v) | 50 mg |
| Chloroform* | 0.2% (v/v) | 0.2 mL |
| Autoclaved ddH2O | n/a | Add up to 100 mL H2O |
| Total | n/a | 100 mL |
*Chloroform is added as a preservative and is optional.
Store TES at 4 °C.
3. Vogel’s minimal medium + sucrose (VMM+S)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 50× Vogel’s solution | 1× | 10 mL |
| Sucrose | 20 g/L | 10 g |
| Agar-agar | 2% (w/v) | 10 g |
| ddH2O | n/a | Add up to 500 mL |
| Total | n/a | 500 mL |
The pH of the minimal medium is about 5.8. No adjustment is necessary.
Autoclave at 112 °C. Pour into flasks or slant agar tubes on a sterile workbench. Allow to cool and set at RT. Store at 4 °C until needed.
4. Vogel’s minimal medium + sorbose-fructose-glucose + cyclosporin A (VMM+SGF+CsA)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 50× Vogel’s solution | 1× | 10 mL |
| Agar-agar | 2% (w/v) | 10 g |
| ddH2O | n/a | Add up to 475 mL |
| SGF* | 5% (v/v) | 25 mL |
| Cyclosporin A (5 mg/mL)* | 5 μg/mL | 500 μL |
| Total | n/a | 500 mL |
*Add after autoclaving.
Autoclave at 121 °C, mix, and pour into 92 mm × 16 mm Petri dishes in a sterile workbench. Allow to cool and set at RT. Store at 4 °C until needed.
5. Sorbose-fructose-glucose solution (SGF)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Sorbose | 20% (w/v) | 20 g |
| Fructose | 0.5% (w/v) | 0.5 g |
| Glucose | 0.5% (w/v) | 0.5 g |
| ddH2O | n/a | Add up to 100 mL |
| Total | n/a | 100 mL |
Autoclave at 112 °C. Let it cool down at RT. Store at 4 °C until needed.
6. Top agar
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 50× Vogel’s solution | 1× | 2 mL |
| Agarose | 1% (w/v) | 1 g |
| ddH2O | n/a | Add up to 95 mL |
| SGF* | 5% (v/v) | 5 mL |
| Cyclosporin A (5 mg/mL)* | 5 μg/mL | 100 μL |
| Total | n/a | 100 mL |
*Add after autoclaving.
Autoclave at 121 °C, add SGF, and keep at 45 °C to avoid solidification.
7. TPS buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Tris-HCl, pH 8.0 | 100 mM | 10 mL |
| 0.5 M EDTA, pH 8.0 | 10 mM | 2 mL |
| KCl | 1 M | 7.55 g |
| ddH2O | n/a | Add up to 100 mL |
| Total | n/a | 100 mL |
Autoclave at 121 °C or filter sterilize. Store at RT.
8. MKM medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Sucrose | 0.5% (w/v) | 2 g |
| 2× SC solution (Recipe 9) | 0.1× | 10 mL |
| 100 mM Na-Iodoacetate solution* | 1 mM | 2 mL |
| Agar-agar | 2% (w/v) | 4 g |
| ddH2O | n/a | Add up to 198 mL |
| Total | n/a | 200 mL |
*Add after autoclaving.
Autoclave at 112 °C. After cooling to 45 °C, add the Na-Iodoacetate and pour into 92 mm × 16 mm Petri dishes in a sterile workbench. Allow to cool and set at RT. Store at 4 °C until needed.
9. 2× Synthetic crossing (SC) solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| KNO3 | 19.8 mM | 400 mg |
| K2HPO4 | 8 mM | 280 mg |
| KH2PO4 | 7.34 mM | 200 mg |
| MgSO4·7H2O | 8 mM | 400 mg |
| NaCl | 3.4 mM | 40 mg |
| CaCl2·2H2O | 1.4 mM | 40 mg |
| Biotin | 0.04 mM | 2 mg |
| TES | 0.02% (v/v) | 40 μL |
| Chloroform* | 0.5% (v/v) | 1 mL |
| Autoclaved ddH2O | n/a | Add up to 200 mL |
| Total | n/a | 200 mL |
*Chloroform is added as a preservative and is optional.
Store at 4 °C in the dark.
10. 1 M Sorbitol + 0.25 % Tween 80
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Sorbitol | 1 M | 18,2 g |
| Tween 80 | 0.25 % (v/v) | 0.25 mL |
| ddH2O | n/a | Add up to 100 mL |
| Total | n/a | 100 mL |
Autoclave at 112 °C. Store at RT.
Laboratory supplies
1. Erlenmeyer Flask, wide neck, 200 mL (Roth, catalog number: X737.1)
2. 1.5 mL Eppendorf tube (Sarstedt, catalog number: 72.690.001)
3. 2 mL Eppendorf tube (Sarstedt, catalog number: 72.695.500)
4. 50 mL Sarstedt tube (Sarstedt, catalog number: 62.547.254)
5. 1,000 μL pipette tips (Sarstedt, catalog number: 70.3050.020)
6. 200 μL pipette tips (Sarstedt, catalog number: 70.3030.020)
7. 10 μL pipette tips (Sarstedt, catalog number: 70.3010)
8. Petri dish, 92 mm × 16 mm (Sarstedt, catalog number: 82.1473.001)
9. Electroporation cuvette, 0.2 cm (Roth, catalog number: PP39.1)
10. Gauze (from medical supplier)
11. Funnel 65 mm (Roth, catalog number: HY46.1)
12. Parafilm M (Roth, catalog number: CNP8.1)
13. Glass test tube with beaded rim, 180 mm × 18 mm (Roth, catalog number: C209.1)
14. Glass test tube with no rim, 130 mm × 16 mm (Roth, catalog number: C188.1)
15. Cellophane made from viscose (any online supplier)
16. Whatman paper 0.35 mm thick (Roth, catalog number: CL66.1)
Equipment
1. PCR machine (e.g., SensoQuest Labcycler, biolab, catalog number: 11-011-103-XXX)
2. Thoma counting chamber (Roth, catalog number: T732.1)
3. Centrifuge (e.g., Beckmann Coulter, model: Allegra X-30R)
4. Electroporation device (e.g., Bio-Rad, model: Genepulser II)
5. Pipettes (Eppendorf)
6. Lancet needle (Roth, catalog number: KY00.1)
7. Inoculation loop (Roth, catalog number: KL99.1)
8. Heating block (e.g., Eppendorf, model: Thermomixer comfort)
9. NanoDrop
Software and datasets
1. CHOP-CHOP tool (https://chopchop.cbu.uib.no/) [15]; online tool used for gRNA design
Procedure
文章信息
稿件历史记录
提交日期: Oct 15, 2025
接收日期: Dec 7, 2025
在线发布日期: Dec 15, 2025
出版日期: Jan 5, 2026
版权信息
© 2026 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Grüttner, S. (2026). Creating Loss-of-Function Mutants of Neurospora crassa Using a Novel CRISPR/Cas9 System. Bio-protocol 16(1): e5562. DOI: 10.21769/BioProtoc.5562.
分类
微生物学 > 微生物遗传学 > CRISPR-Cas9
分子生物学 > DNA > 诱/突变
微生物学 > 微生物遗传学 > 基因组编辑
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link